Virus-Encoded Complement Regulators: Current Status
- PMID: 33573085
- PMCID: PMC7912105
- DOI: 10.3390/v13020208
Virus-Encoded Complement Regulators: Current Status
Abstract
Viruses require a host for replication and survival and hence are subjected to host immunological pressures. The complement system, a crucial first response of the host immune system, is effective in targeting viruses and virus-infected cells, and boosting the antiviral innate and acquired immune responses. Thus, the system imposes a strong selection pressure on viruses. Consequently, viruses have evolved multiple countermeasures against host complement. A major mechanism employed by viruses to subvert the complement system is encoding proteins that target complement. Since viruses have limited genome size, most of these proteins are multifunctional in nature. In this review, we provide up to date information on the structure and complement regulatory functions of various viral proteins.
Keywords: CD59; Herpesvirus; Poxvirus; RCA; complement; innate immunity; pathogenesis; viral RCA; viral immune evasion.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures
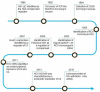
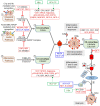
References
-
- Bordet J., Gengou O. Sur l’Existences de Substances Sensibilisatrices dans la Plupart des Serums Anti-Microbiens. Ann. I'Institut Pasteur. 1901;15:289–302.
-
- Ember J.A., Jagels M.A., Hugli T.E. Characterization of Complement Anaphylatoxins and Their Biological Responses. In: Volanakis J.E., Frank M.M., editors. The Human Complement System in Health and Disease. Marcel Dekker, Inc.; New York, NY, USA: 1998. Chapter 11.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

