Highly Pathogenic Avian Influenza Viruses at the Wild-Domestic Bird Interface in Europe: Future Directions for Research and Surveillance
- PMID: 33573231
- PMCID: PMC7912471
- DOI: 10.3390/v13020212
Highly Pathogenic Avian Influenza Viruses at the Wild-Domestic Bird Interface in Europe: Future Directions for Research and Surveillance
Abstract
Highly pathogenic avian influenza (HPAI) outbreaks in wild birds and poultry are no longer a rare phenomenon in Europe. In the past 15 years, HPAI outbreaks-in particular those caused by H5 viruses derived from the A/Goose/Guangdong/1/1996 lineage that emerged in southeast Asia in 1996-have been occuring with increasing frequency in Europe. Between 2005 and 2020, at least ten HPAI H5 incursions were identified in Europe resulting in mass mortalities among poultry and wild birds. Until 2009, the HPAI H5 virus outbreaks in Europe were caused by HPAI H5N1 clade 2.2 viruses, while from 2014 onwards HPAI H5 clade 2.3.4.4 viruses dominated outbreaks, with abundant genetic reassortments yielding subtypes H5N1, H5N2, H5N3, H5N4, H5N5, H5N6 and H5N8. The majority of HPAI H5 virus detections in wild and domestic birds within Europe coincide with southwest/westward fall migration and large local waterbird aggregations during wintering. In this review we provide an overview of HPAI H5 virus epidemiology, ecology and evolution at the interface between poultry and wild birds based on 15 years of avian influenza virus surveillance in Europe, and assess future directions for HPAI virus research and surveillance, including the integration of whole genome sequencing, host identification and avian ecology into risk-based surveillance and analyses.
Keywords: aquatic birds; avian influenza; ecology; epidemiology; evolution; highly pathogenic avian influenza; influenza subtypes; influenza surveillance; migratory birds; migratory flyways.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures
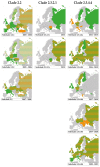
References
-
- FAOSTAT Food and Agriculture Data of the FAO–Food and Agriculture Organization of the United Nations. [(accessed on 17 December 2020)]; Available online: http://www.fao.org/faostat/en/#home.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

