Bile Acids and Microbiota: Multifaceted and Versatile Regulators of the Liver-Gut Axis
- PMID: 33573273
- PMCID: PMC7866539
- DOI: 10.3390/ijms22031397
Bile Acids and Microbiota: Multifaceted and Versatile Regulators of the Liver-Gut Axis
Abstract
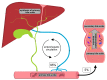
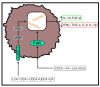
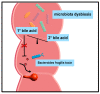
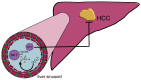
After their synthesis from cholesterol in hepatic tissues, bile acids (BAs) are secreted into the intestinal lumen. Most BAs are subsequently re-absorbed in the terminal ileum and are transported back for recycling to the liver. Some of them, however, reach the colon and change their physicochemical properties upon modification by gut bacteria, and vice versa, BAs also shape the composition and function of the intestinal microbiota. This mutual interplay of both BAs and gut microbiota regulates many physiological processes, including the lipid, carbohydrate and energy metabolism of the host. Emerging evidence also implies an important role of this enterohepatic BA circuit in shaping mucosal colonization resistance as well as local and distant immune responses, tissue physiology and carcinogenesis. Subsequently, disrupted interactions of gut bacteria and BAs are associated with many disorders as diverse as Clostridioides difficile or Salmonella Typhimurium infection, inflammatory bowel disease, type 1 diabetes, asthma, metabolic syndrome, obesity, Parkinson's disease, schizophrenia and epilepsy. As we cannot address all of these interesting underlying pathophysiologic mechanisms here, we summarize the current knowledge about the physiologic and pathogenic interplay of local site microbiota and the enterohepatic BA metabolism using a few selected examples of liver and gut diseases.
Keywords: bile acids; enterohepatic recirculation; host–microbe interactions; intestinal microbiota; microbial metabolism.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

