Communicating clocks shape circadian homeostasis
- PMID: 33574181
- PMCID: PMC8123919
- DOI: 10.1126/science.abd0951
Communicating clocks shape circadian homeostasis
Abstract
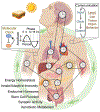
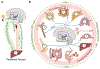
Circadian clocks temporally coordinate physiology and align it with geophysical time, which enables diverse life-forms to anticipate daily environmental cycles. In complex organisms, clock function originates from the molecular oscillator within each cell and builds upward anatomically into an organism-wide system. Recent advances have transformed our understanding of how clocks are connected to achieve coherence across tissues. Circadian misalignment, often imposed in modern society, disrupts coordination among clocks and has been linked to diseases ranging from metabolic syndrome to cancer. Thus, uncovering the physiological circuits whereby biological clocks achieve coherence will inform on both challenges and opportunities in human health.
Copyright © 2021 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
Figures




References
-
- Whitmore D, Foulkes NS, Sassone-Corsi P, Light acts directly on organs and cells in culture to set the vertebrate circadian clock. Nature 404, 87–91 (2000). - PubMed
-
- Moore RY, Eichler VB, Loss of a circadian adrenal corticosterone rhythm following suprachiasmatic lesions in the rat. Brain Res 42, 201–206 (1972). - PubMed
-
- Schibler U, Sassone-Corsi P, A web of circadian pacemakers. Cell 111, 919–922 (2002). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

