Cellular transcriptomics reveals evolutionary identities of songbird vocal circuits
- PMID: 33574185
- PMCID: PMC8136249
- DOI: 10.1126/science.abd9704
Cellular transcriptomics reveals evolutionary identities of songbird vocal circuits
Abstract
Birds display advanced behaviors, including vocal learning and problem-solving, yet lack a layered neocortex, a structure associated with complex behavior in mammals. To determine whether these behavioral similarities result from shared or distinct neural circuits, we used single-cell RNA sequencing to characterize the neuronal repertoire of the songbird song motor pathway. Glutamatergic vocal neurons had considerable transcriptional similarity to neocortical projection neurons; however, they displayed regulatory gene expression patterns more closely related to neurons in the ventral pallium. Moreover, while γ-aminobutyric acid-releasing neurons in this pathway appeared homologous to those in mammals and other amniotes, the most abundant avian class is largely absent in the neocortex. These data suggest that songbird vocal circuits and the mammalian neocortex have distinct developmental origins yet contain transcriptionally similar neurons.
Copyright © 2021 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
Figures


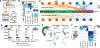


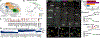
Comment in
-
Different origins for similar brain circuits.Science. 2021 Feb 12;371(6530):676-677. doi: 10.1126/science.abf9551. Science. 2021. PMID: 33574199 No abstract available.
References
-
- Striedter GF, Northcutt RG, Brains Through Time: A Natural History of Vertebrates (Oxford Univ. Press, 2020).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

