Human myelomeningocele risk and ultra-rare deleterious variants in genes associated with cilium, WNT-signaling, ECM, cytoskeleton and cell migration
- PMID: 33574475
- PMCID: PMC7878900
- DOI: 10.1038/s41598-021-83058-7
Human myelomeningocele risk and ultra-rare deleterious variants in genes associated with cilium, WNT-signaling, ECM, cytoskeleton and cell migration
Abstract
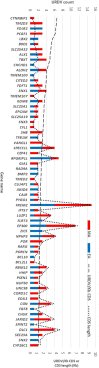
Myelomeningocele (MMC) affects one in 1000 newborns annually worldwide and each surviving child faces tremendous lifetime medical and caregiving burdens. Both genetic and environmental factors contribute to disease risk but the mechanism is unclear. This study examined 506 MMC subjects for ultra-rare deleterious variants (URDVs, absent in gnomAD v2.1.1 controls that have Combined Annotation Dependent Depletion score ≥ 20) in candidate genes either known to cause abnormal neural tube closure in animals or previously associated with human MMC in the current study cohort. Approximately 70% of the study subjects carried one to nine URDVs among 302 candidate genes. Half of the study subjects carried heterozygous URDVs in multiple genes involved in the structure and/or function of cilium, cytoskeleton, extracellular matrix, WNT signaling, and/or cell migration. Another 20% of the study subjects carried heterozygous URDVs in candidate genes associated with gene transcription regulation, folate metabolism, or glucose metabolism. Presence of URDVs in the candidate genes involving these biological function groups may elevate the risk of developing myelomeningocele in the study cohort.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

