The gut microbiome modulates the protective association between a Mediterranean diet and cardiometabolic disease risk
- PMID: 33574608
- PMCID: PMC8186452
- DOI: 10.1038/s41591-020-01223-3
The gut microbiome modulates the protective association between a Mediterranean diet and cardiometabolic disease risk
Abstract
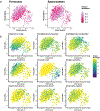
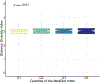
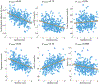
To address how the microbiome might modify the interaction between diet and cardiometabolic health, we analyzed longitudinal microbiome data from 307 male participants in the Health Professionals Follow-Up Study, together with long-term dietary information and measurements of biomarkers of glucose homeostasis, lipid metabolism and inflammation from blood samples. Here, we demonstrate that a healthy Mediterranean-style dietary pattern is associated with specific functional and taxonomic components of the gut microbiome, and that its protective associations with cardiometabolic health vary depending on microbial composition. In particular, the protective association between adherence to the Mediterranean diet and cardiometabolic disease risk was significantly stronger among participants with decreased abundance of Prevotella copri. Our findings advance the concept of precision nutrition and have the potential to inform more effective and precise dietary approaches for the prevention of cardiometabolic disease mediated through alterations in the gut microbiome.
Figures
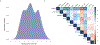





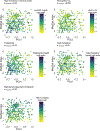


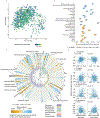


Comment in
-
Modulating the gut microbiota with dietary interventions to protect against cardiometabolic disease.Nat Rev Cardiol. 2021 May;18(5):305. doi: 10.1038/s41569-021-00537-0. Nat Rev Cardiol. 2021. PMID: 33658636 No abstract available.
-
Modulating the gut microbiome with dietary interventions to reduce cardiometabolic disease risk.Eur Heart J. 2021 Jun 7;42(22):2152-2153. doi: 10.1093/eurheartj/ehab261. Eur Heart J. 2021. PMID: 34097727 No abstract available.
References
-
- DALYs G.B.D. & Collaborators H. Global, regional, and national disability-adjusted life-years (DALYs) for 359 diseases and injuries and healthy life expectancy (HALE) for 195 countries and territories, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet 392, 1859–1922 (2018). - PMC - PubMed
-
- Kurilshikov A, et al. Gut Microbial Associations to Plasma Metabolites Linked to Cardiovascular Phenotypes and Risk. Circ Res 124, 1808–1820 (2019). - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
- P30DK046200/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- P30 DK043351/DK/NIDDK NIH HHS/United States
- U01 CA152904/CA/NCI NIH HHS/United States
- U01 CA167552/CA/NCI NIH HHS/United States
- R00DK119412/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- R01 HL035464/HL/NHLBI NIH HHS/United States
- R01 HL060712/HL/NHLBI NIH HHS/United States
- R01CA202704/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- U01CA167552/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- R00 DK119412/DK/NIDDK NIH HHS/United States
- R01HL035464/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- K99 DK119412/DK/NIDDK NIH HHS/United States
- R01 CA202704/CA/NCI NIH HHS/United States
- K24DK098311/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- P30 DK046200/DK/NIDDK NIH HHS/United States
- K24 DK098311/DK/NIDDK NIH HHS/United States
- K23 DK125838/DK/NIDDK NIH HHS/United States
- U54 DE023798/DE/NIDCR NIH HHS/United States
- R01HL060712/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- U54DE023798/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- U01CA152904/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
LinkOut - more resources
Full Text Sources
Other Literature Sources

