Comparative Virulence and Genomic Analysis of Streptococcus suis Isolates
- PMID: 33574803
- PMCID: PMC7870872
- DOI: 10.3389/fmicb.2020.620843
Comparative Virulence and Genomic Analysis of Streptococcus suis Isolates
Abstract
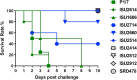
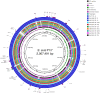
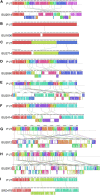
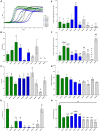
Streptococcus suis is a zoonotic bacterial swine pathogen causing substantial economic and health burdens to the pork industry. Mechanisms used by S. suis to colonize and cause disease remain unknown and vaccines and/or intervention strategies currently do not exist. Studies addressing virulence mechanisms used by S. suis have been complicated because different isolates can cause a spectrum of disease outcomes ranging from lethal systemic disease to asymptomatic carriage. The objectives of this study were to evaluate the virulence capacity of nine United States S. suis isolates following intranasal challenge in swine and then perform comparative genomic analyses to identify genomic attributes associated with swine-virulent phenotypes. No correlation was found between the capacity to cause disease in swine and the functional characteristics of genome size, serotype, sequence type (ST), or in vitro virulence-associated phenotypes. A search for orthologs found in highly virulent isolates and not found in non-virulent isolates revealed numerous predicted protein coding sequences specific to each category. While none of these predicted protein coding sequences have been previously characterized as potential virulence factors, this analysis does provide a reliable one-to-one assignment of specific genes of interest that could prove useful in future allelic replacement and/or functional genomic studies. Collectively, this report provides a framework for future allelic replacement and/or functional genomic studies investigating genetic characteristics underlying the spectrum of disease outcomes caused by S. suis isolates.
Keywords: Streptococcus suis; antimicrobial resistance; comparative genomics; mobile genetic elements; swine; virulence; whole-genome sequencing.
Copyright © 2021 Nicholson, Waack, Anderson, Bayles, Zaia, Goertz, Eppinger, Hau, Brockmeier and Shore.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
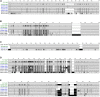
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

