Degradation of BRD4 - a promising treatment approach not only for hematologic but also for solid cancer
- PMID: 33575085
- PMCID: PMC7868748
Degradation of BRD4 - a promising treatment approach not only for hematologic but also for solid cancer
Abstract
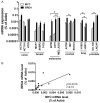
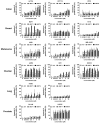
Bromodomain (BRD) and extra-terminal (BET) proteins are epigenetic readers that regulate gene expression and promote cancer evolution. Pharmacological inactivation of BRD4 has recently been introduced as a promising anti-neoplastic approach that targets MYC oncogene expression. However, resistance against BRD4-targeting drugs has been described. We compared the efficacy of the small-molecule-type BET BRD inhibitor JQ1 with the recently developed BET protein degraders dBET1 and dBET6 in colon, breast, melanoma, ovarian, lung and prostate cancer cell lines. As determined by qPCR, all BRD4 targeting drugs dose-dependently decreased MYC expression, with dBET6 introducing the strongest downregulation of MYC. This correlated with the anti-proliferative activity of these drugs, which was at least one order of magnitude higher for dBET6 (IC50 0.001-0.5 µM) than for dBET1 or JQ1 (IC50 0.5-5 µM). Interestingly, when combined with commonly used cytotoxic therapeutics, dBET6 was found to promote anti-neoplastic effects and to counteract chemoresistance in most cancer cell lines. Moreover, JQ1 and both BET degraders strongly downregulated baseline and interferon-gamma induced expression of the immune checkpoint molecule PD-L1 in all cancer cell lines. Together, our data suggest that dBET6 outperforms first-generation BRD4 targeting drugs like dBET1 and JQ1, and decreases chemoresistance and immune resistance of cancer.
Keywords: BET degrader; MYC; PD-L1; dBET6; solid tumor.
AJCR Copyright © 2021.
Conflict of interest statement
None.
Figures




References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
