Identifying microbial species by single-molecule DNA optical mapping and resampling statistics
- PMID: 33575560
- PMCID: PMC7671359
- DOI: 10.1093/nargab/lqz007
Identifying microbial species by single-molecule DNA optical mapping and resampling statistics
Abstract
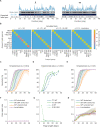
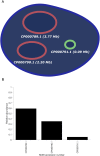
Single-molecule DNA mapping has the potential to serve as a powerful complement to high-throughput sequencing in metagenomic analysis. Offering longer read lengths and forgoing the need for complex library preparation and amplification, mapping stands to provide an unbiased view into the composition of complex viromes and/or microbiomes. To fully enable mapping-based metagenomics, sensitivity and specificity of DNA map analysis and identification need to be improved. Using detailed simulations and experimental data, we first demonstrate how fluorescence imaging of surface stretched, sequence specifically labeled DNA fragments can yield highly sensitive identification of targets. Second, a new analysis technique is introduced to increase specificity of the analysis, allowing even closely related species to be resolved. Third, we show how an increase in resolution improves sensitivity. Finally, we demonstrate that these methods are capable of identifying species with long genomes such as bacteria with high sensitivity.
© The Author(s) 2019. Published by Oxford University Press on behalf of NAR Genomics and Bioinformatics.
Figures



References
-
- White R.A., Callister S.J., Moore R.J., Baker E.S., Jansson J.K., White R. III, J Callister S., Moore R.J., Baker E.S., Jansson J.K. et al. .. The past, present and future of microbiome analyses. Nat. Protoc. 2016; 11:2049–2053.
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

