Unraveling heteroplasmy patterns with NOVOPlasty
- PMID: 33575563
- PMCID: PMC7671380
- DOI: 10.1093/nargab/lqz011
Unraveling heteroplasmy patterns with NOVOPlasty
Abstract
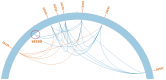
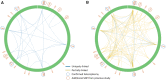
Heteroplasmy, the existence of multiple mitochondrial haplotypes within an individual, has been studied across different scientific fields. Mitochondrial genome polymorphisms have been linked to multiple severe disorders and are of interest to evolutionary studies and forensic science. Before the development of massive parallel sequencing (MPS), most studies of mitochondrial genome variation were limited to short fragments and to heteroplasmic variants associated with a relatively high frequency (>10%). By utilizing ultra-deep sequencing, it has now become possible to uncover previously undiscovered patterns of intra-individual polymorphisms. Despite these technological advances, it is still challenging to determine the origin of the observed intra-individual polymorphisms. We therefore developed a new method that not only detects intra-individual polymorphisms within mitochondrial and chloroplast genomes more accurately, but also looks for linkage among polymorphic sites by assembling the sequence around each detected polymorphic site. Our benchmark study shows that this method is capable of detecting heteroplasmy more accurately than any method previously available and is the first tool that is able to completely or partially reconstruct the sequence for each mitochondrial haplotype (allele). The method is implemented in our open source software NOVOPlasty that can be downloaded at https://github.com/ndierckx/NOVOPlasty.
© The Author(s) 2019. Published by Oxford University Press on behalf of NAR Genomics and Bioinformatics.
Figures

References
-
- Kvist L., Martens J., Nazarenko A.A., Orell M.. Paternal leakage of mitochondrial DNA in the great tit (Parus major). Mol. Biol. Evol. 2003; 20:243–247. - PubMed
LinkOut - more resources
Full Text Sources

