Genetically regulated expression underlies cellular sensitivity to chemotherapy in diverse populations
- PMID: 33575800
- PMCID: PMC8248963
- DOI: 10.1093/hmg/ddab029
Genetically regulated expression underlies cellular sensitivity to chemotherapy in diverse populations
Abstract
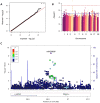
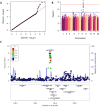
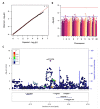
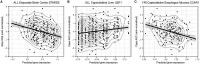
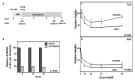
Most cancer chemotherapeutic agents are ineffective in a subset of patients; thus, it is important to consider the role of genetic variation in drug response. Lymphoblastoid cell lines (LCLs) in 1000 Genomes Project populations of diverse ancestries are a useful model for determining how genetic factors impact the variation in cytotoxicity. In our study, LCLs from three 1000 Genomes Project populations of diverse ancestries were previously treated with increasing concentrations of eight chemotherapeutic drugs, and cell growth inhibition was measured at each dose with half-maximal inhibitory concentration (IC50) or area under the dose-response curve (AUC) as our phenotype for each drug. We conducted both genome-wide association studies (GWAS) and transcriptome-wide association studies (TWAS) within and across ancestral populations. We identified four unique loci in GWAS and three genes in TWAS to be significantly associated with the chemotherapy-induced cytotoxicity within and across ancestral populations. In the etoposide TWAS, increased STARD5 predicted expression associated with decreased etoposide IC50 (P = 8.5 × 10-8). Functional studies in A549, a lung cancer cell line, revealed that knockdown of STARD5 expression resulted in the decreased sensitivity to etoposide following exposure for 72 (P = 0.033) and 96 h (P = 0.0001). By identifying loci and genes associated with cytotoxicity across ancestral populations, we strive to understand the genetic factors impacting the effectiveness of chemotherapy drugs and to contribute to the development of future cancer treatment.
© The Author(s) 2021. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures
Similar articles
-
Radiation pharmacogenomics: a genome-wide association approach to identify radiation response biomarkers using human lymphoblastoid cell lines.Genome Res. 2010 Nov;20(11):1482-92. doi: 10.1101/gr.107672.110. Epub 2010 Oct 5. Genome Res. 2010. PMID: 20923822 Free PMC article.
-
A genome-wide approach to identify genetic variants that contribute to etoposide-induced cytotoxicity.Proc Natl Acad Sci U S A. 2007 Jun 5;104(23):9758-63. doi: 10.1073/pnas.0703736104. Epub 2007 May 30. Proc Natl Acad Sci U S A. 2007. PMID: 17537913 Free PMC article.
-
Heritable and non-genetic factors as variables of pharmacologic phenotypes in lymphoblastoid cell lines.Pharmacogenomics J. 2010 Dec;10(6):505-12. doi: 10.1038/tpj.2010.3. Epub 2010 Feb 9. Pharmacogenomics J. 2010. PMID: 20142840 Free PMC article.
-
Lymphoblastoid cell lines in pharmacogenomic discovery and clinical translation.Pharmacogenomics. 2012 Jan;13(1):55-70. doi: 10.2217/pgs.11.121. Pharmacogenomics. 2012. PMID: 22176622 Free PMC article. Review.
-
From GWAS to Gene: Transcriptome-Wide Association Studies and Other Methods to Functionally Understand GWAS Discoveries.Front Genet. 2021 Sep 30;12:713230. doi: 10.3389/fgene.2021.713230. eCollection 2021. Front Genet. 2021. PMID: 34659337 Free PMC article. Review.
Cited by
-
Analysis of immune cell infiltration in the tumor microenvironment of cervical cancer and its impact on immunotherapy.Front Oncol. 2025 Jul 3;15:1608597. doi: 10.3389/fonc.2025.1608597. eCollection 2025. Front Oncol. 2025. PMID: 40678068 Free PMC article.
-
FinnGen provides genetic insights from a well-phenotyped isolated population.Nature. 2023 Jan;613(7944):508-518. doi: 10.1038/s41586-022-05473-8. Epub 2023 Jan 18. Nature. 2023. PMID: 36653562 Free PMC article.
-
Protein prediction for trait mapping in diverse populations.PLoS One. 2022 Feb 24;17(2):e0264341. doi: 10.1371/journal.pone.0264341. eCollection 2022. PLoS One. 2022. PMID: 35202437 Free PMC article.
-
Eleven metabolism‑related genes composed of Stard5 predict prognosis and contribute to EMT phenotype in HCC.Cancer Cell Int. 2023 Nov 17;23(1):277. doi: 10.1186/s12935-023-03097-0. Cancer Cell Int. 2023. PMID: 37978523 Free PMC article.
-
AteMeVs: An R package for the estimation of the average treatment effect with measurement error and variable selection for confounders.PLoS One. 2024 Sep 30;19(9):e0296951. doi: 10.1371/journal.pone.0296951. eCollection 2024. PLoS One. 2024. PMID: 39348368 Free PMC article.
References
-
- Torre, L.A., Siegel, R.L., Ward, E.M. and Jemal, A. (2016) Global cancer incidence and mortality rates and trends—an update. Cancer Epidemiol. Biomark. Prev., 25, 16–27. - PubMed
-
- Galmarini, D., Galmarini, C.M. and Galmarini, F.C. (2012) Cancer chemotherapy: a critical analysis of its 60 years of history. Crit. Rev. Oncol. Hematol., 84, 181–199. - PubMed
-
- Marin, J.J.G., Romero, M.R., Blazquez, A.G., Herraez, E., Keck, E. and Briz, O. (2009) Importance and limitations of chemotherapy among the available treatments for gastrointestinal tumours. Anti Cancer Agents Med. Chem., 9, 162–184. - PubMed
-
- Roy, P.S. and Saikia, B.J. (2016) Cancer and cure: a critical analysis. Indian J. Cancer, 53, 441–442. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

