Regulatory T cells promote glioma cell stemness through TGF-β-NF-κB-IL6-STAT3 signaling
- PMID: 33576874
- PMCID: PMC8360896
- DOI: 10.1007/s00262-021-02872-0
Regulatory T cells promote glioma cell stemness through TGF-β-NF-κB-IL6-STAT3 signaling
Abstract
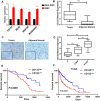
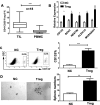
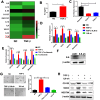
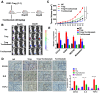
Glioma stem cells (GSCs) contribute to the malignant growth of glioma, but little is known about the interaction between GSCs and tumor microenvironment. Here, we found that intense infiltration of regulatory T cells (Tregs) facilitated the qualities of GSCs through TGF-β secretion that helped coordinately tumor growth. Mechanistic investigations indicated that TGF-β acted on cancer cells to induce the core cancer stem cell-related genes CD133, SOX2, NESTIN, MUSASHI1 and ALDH1A expression and spheres formation via NF-κB-IL6-STAT3 signaling pathway, resulting in the increased cancer stemness and tumorigenic potential. Furthermore, Tregs promoted glioma tumor growth, and this effect could be abrogated with blockade of IL6 receptor by tocilizumab which also demonstrated certain level of therapeutic efficacy in xenograft model. Additionally, expression levels of CD133, IL6 and TGF-β were found to serve as prognosis markers of glioma patients. Collectively, our findings reveal a new immune-associated mechanism underlying Tregs-induced GSCs. Moreover, efforts to target this network may be an effective strategy for treating glioma.
Keywords: Glioma; Glioma stem cell; TGF-β; Tocilizumab; Tregs.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no conflict of interests.
Figures


References
-
- Brada M, Yung WK. Clinical trial end points in malignant glioma: need for effective trial design strategy. Semin Oncol. 2000;27:11–19. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

