MetNet: A two-level approach to reconstructing and comparing metabolic networks
- PMID: 33577575
- PMCID: PMC7880445
- DOI: 10.1371/journal.pone.0246962
MetNet: A two-level approach to reconstructing and comparing metabolic networks
Abstract
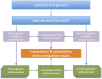
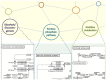
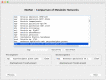
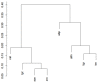
Metabolic pathway comparison and interaction between different species can detect important information for drug engineering and medical science. In the literature, proposals for reconstructing and comparing metabolic networks present two main problems: network reconstruction requires usually human intervention to integrate information from different sources and, in metabolic comparison, the size of the networks leads to a challenging computational problem. We propose to automatically reconstruct a metabolic network on the basis of KEGG database information. Our proposal relies on a two-level representation of the huge metabolic network: the first level is graph-based and depicts pathways as nodes and relations between pathways as edges; the second level represents each metabolic pathway in terms of its reactions content. The two-level representation complies with the KEGG database, which decomposes the metabolism of all the different organisms into "reference" pathways in a standardised way. On the basis of this two-level representation, we introduce some similarity measures for both levels. They allow for both a local comparison, pathway by pathway, and a global comparison of the entire metabolism. We developed a tool, MetNet, that implements the proposed methodology. MetNet makes it possible to automatically reconstruct the metabolic network of two organisms selected in KEGG and to compare their two networks both quantitatively and visually. We validate our methodology by presenting some experiments performed with MetNet.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Metabolomics analysis: Finding out metabolic building blocks.PLoS One. 2017 May 11;12(5):e0177031. doi: 10.1371/journal.pone.0177031. eCollection 2017. PLoS One. 2017. PMID: 28493998 Free PMC article.
-
Computational reconstruction of metabolic networks from KEGG.Methods Mol Biol. 2013;930:235-49. doi: 10.1007/978-1-62703-059-5_10. Methods Mol Biol. 2013. PMID: 23086844
-
Putting The Plant Metabolic Network pathway databases to work: going offline to gain new capabilities.Methods Mol Biol. 2014;1083:151-71. doi: 10.1007/978-1-62703-661-0_10. Methods Mol Biol. 2014. PMID: 24218215
-
Prediction of metabolic pathways from genome-scale metabolic networks.Biosystems. 2011 Aug;105(2):109-21. doi: 10.1016/j.biosystems.2011.05.004. Epub 2011 May 27. Biosystems. 2011. PMID: 21645586 Review.
-
Metabolomics assisted metabolic network modeling and network wide analysis of metabolites in microbiology.Crit Rev Biotechnol. 2018 Nov;38(7):1106-1120. doi: 10.1080/07388551.2018.1462141. Epub 2018 Apr 22. Crit Rev Biotechnol. 2018. PMID: 29683004 Review.
Cited by
-
Exploring the expressiveness of abstract metabolic networks.PLoS One. 2023 Feb 9;18(2):e0281047. doi: 10.1371/journal.pone.0281047. eCollection 2023. PLoS One. 2023. PMID: 36758030 Free PMC article.
-
Mitochondrial Metabolomics in Cancer: Mass Spectrometry-Based Approaches for Metabolic Rewiring Analysis and Therapeutic Discovery.Metabolites. 2025 Jul 31;15(8):513. doi: 10.3390/metabo15080513. Metabolites. 2025. PMID: 40863132 Free PMC article. Review.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

