Leveraging the antiviral type I interferon system as a first line of defense against SARS-CoV-2 pathogenicity
- PMID: 33577760
- PMCID: PMC7846242
- DOI: 10.1016/j.immuni.2021.01.017
Leveraging the antiviral type I interferon system as a first line of defense against SARS-CoV-2 pathogenicity
Abstract
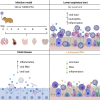
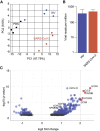
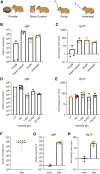
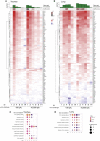
The emergence and spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has resulted in significant global morbidity, mortality, and societal disruption. A better understanding of virus-host interactions may potentiate therapeutic insights toward limiting this infection. Here we investigated the dynamics of the systemic response to SARS-CoV-2 in hamsters by histological analysis and transcriptional profiling. Infection resulted in consistently high levels of virus in the upper and lower respiratory tracts and sporadic occurrence in other distal tissues. A longitudinal cohort revealed a wave of inflammation, including a type I interferon (IFN-I) response, that was evident in all tissues regardless of viral presence but was insufficient to prevent disease progression. Bolstering the antiviral response with intranasal administration of recombinant IFN-I reduced viral disease, prevented transmission, and lowered inflammation in vivo. This study defines the systemic host response to SARS-CoV-2 infection and supports use of intranasal IFN-I as an effective means of early treatment.
Keywords: COVID-19; IFN-I; cytokine; hamster; intranasal; mRNA-seq; pandemic; prophylactic; therapeutic; transcriptomics.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



References
-
- Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D.A., Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. - PubMed
-
- Bastard P., Rosen L.B., Zhang Q., Michailidis E., Hoffmann H.H., Zhang Y., Dorgham K., Philippot Q., Rosain J., Béziat V., et al. HGID Lab. NIAID-USUHS Immune Response to COVID Group. COVID Clinicians. COVID-STORM Clinicians. Imagine COVID Group. French COVID Cohort Study Group. Milieu Intérieur Consortium. CoV-Contact Cohort. Amsterdam UMC Covid-19 Biobank. COVID Human Genetic Effort Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370:eabd4585. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

