Two-component spike nanoparticle vaccine protects macaques from SARS-CoV-2 infection
- PMID: 33577765
- PMCID: PMC7834972
- DOI: 10.1016/j.cell.2021.01.035
Two-component spike nanoparticle vaccine protects macaques from SARS-CoV-2 infection
Abstract
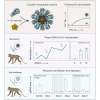
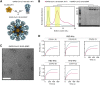
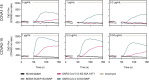
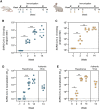
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic is continuing to disrupt personal lives, global healthcare systems, and economies. Hence, there is an urgent need for a vaccine that prevents viral infection, transmission, and disease. Here, we present a two-component protein-based nanoparticle vaccine that displays multiple copies of the SARS-CoV-2 spike protein. Immunization studies show that this vaccine induces potent neutralizing antibody responses in mice, rabbits, and cynomolgus macaques. The vaccine-induced immunity protects macaques against a high-dose challenge, resulting in strongly reduced viral infection and replication in the upper and lower airways. These nanoparticles are a promising vaccine candidate to curtail the SARS-CoV-2 pandemic.
Keywords: B cells; COVID-19; SARS-CoV-2; antibodies; immunity; macaques; nanoparticles; protection; vaccine.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests N.P.K. is a co-founder, shareholder, and chair of the scientific advisory board of Icosavax. The remaining authors declare no competing interests. Amsterdam UMC has filed a patent application concerning the SARS-CoV-2 mAbs used here (Brouwer et al., 2020). N.P.K. has a nonprovisional US patent (no. 14/930,792) related to I53-50 (Bale et al., 2016).
Figures

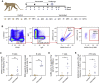
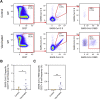
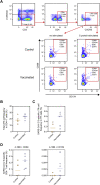
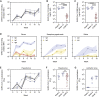



References
-
- Addetia A., Crawford K.H.D., Dingens A., Zhu H., Roychoudhury P., Huang M.L., Jerome K.R., Bloom J.D., Greninger A.L. Neutralizing antibodies correlate with protection from SARS-CoV-2 in humans during a fishery vessel outbreak with high attack rate. J. Clin. Microbiol. 2020;58:e02107–e02120. - PMC - PubMed
-
- Amanat F., Strohmeier S., Rathnasinghe R., Schotsaert M., Coughlan L., García-Sastre A., Krammer F. Introduction of two prolines and removal of the polybasic cleavage site leads to optimal efficacy of a recombinant spike based SARS-CoV-2 vaccine in the mouse model. bioRxiv. 2020 doi: 10.1101/2020.09.16.300970. - DOI - PMC - PubMed
-
- Bachmann M.F., Jennings G.T. Vaccine delivery: a matter of size, geometry, kinetics and molecular patterns. Nat. Rev. Immunol. 2010;10:787–796. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

