Detecting circRNA in purified spliceosomal P complex
- PMID: 33577981
- PMCID: PMC8352997
- DOI: 10.1016/j.ymeth.2021.02.002
Detecting circRNA in purified spliceosomal P complex
Abstract
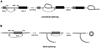
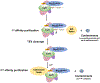
Circular RNAs (circRNAs) generated from back-splicing of exons have been found in a wide range of eukaryotic species and exert a variety of biological functions. Unlike canonical splicing, the mechanism of back-splicing has long remained elusive. We recently determined the cryo-EM structure of the yeast spliceosomal E complex assembled on introns, leading us to hypothesize that the same E complex can assemble across an exon forming the exon-definition complex. This complex, when assembled on long exons, goes through the splicing cycle and catalyzes back-splicing to generate circRNAs. Supporting this hypothesis, we purified the yeast post-catalytic spliceosomal P complex (the best complex in the splicing cycle to trap splicing products and intermediates) and detected canonical and back-splicing products as well as splicing intermediates. Here we describe in detail this procedure, which may be applied to other organisms to facilitate research on the biogenesis and regulation of circRNA.
Keywords: Back-splicing; CircRNA; P complex; Spliceosome.
Copyright © 2021 Elsevier Inc. All rights reserved.
Figures


References
-
- Cocquerelle C, Mascrez B, Hetuin D, Bailleul B, Mis-splicing yields circular RNA molecules, FASEB J 7(1) (1993) 155–60. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

