Peptidomics-Driven Strategy Reveals Peptides and Predicted Proteases Associated With Oral Cancer Prognosis
- PMID: 33578082
- PMCID: PMC7950089
- DOI: 10.1074/mcp.RA120.002227
Peptidomics-Driven Strategy Reveals Peptides and Predicted Proteases Associated With Oral Cancer Prognosis
Abstract
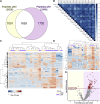
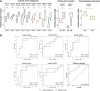
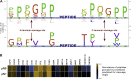
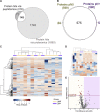
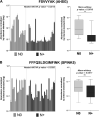
Protease activity has been associated with pathological processes that can lead to cancer development and progression. However, understanding the pathological unbalance in proteolysis is challenging because changes can occur simultaneously at protease, their inhibitor, and substrate levels. Here, we present a pipeline that combines peptidomics, proteomics, and peptidase predictions for studying proteolytic events in the saliva of 79 patients and their association with oral squamous cell carcinoma (OSCC) prognosis. Our findings revealed differences in the saliva peptidome of patients with (pN+) or without (pN0) lymph-node metastasis and delivered a panel of ten endogenous peptides correlated with poor prognostic factors plus five molecules able to classify pN0 and pN+ patients (area under the receiver operating characteristic curve > 0.85). In addition, endopeptidases and exopeptidases putatively implicated in the processing of differential peptides were investigated using cancer tissue gene expression data from public repositories, reinforcing their association with poorer survival rates and prognosis in oral cancer. The dynamics of the OSCC-related proteolysis were further explored via the proteomic profiling of saliva. This revealed that peptidase/endopeptidase inhibitors exhibited reduced levels in the saliva of pN+ patients, as confirmed by selected reaction monitoring-mass spectrometry, while minor changes were detected in the level of saliva proteases. Taken together, our results indicated that proteolytic activity is accentuated in the saliva of patients with OSCC and lymph-node metastasis and, at least in part, is modulated by reduced levels of salivary peptidase inhibitors. Therefore, this integrated pipeline provided better comprehension and discovery of molecular features with implications in the oral cancer metastasis prognosis.
Keywords: head and neck squamous cell carcinoma; peptidomics; proteolysis; saliva.
Copyright © 2020 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare no competing interests.
Figures





References
-
- Doucet A., Butler G.S., Rodríguez D., Prudova A., Overall C.M. Metadegradomics. Mol. Cell. Proteomics. 2008;7:1925–1951. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

