Usp11 controls cortical neurogenesis and neuronal migration through Sox11 stabilization
- PMID: 33579706
- PMCID: PMC7880594
- DOI: 10.1126/sciadv.abc6093
Usp11 controls cortical neurogenesis and neuronal migration through Sox11 stabilization
Abstract
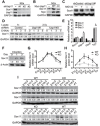
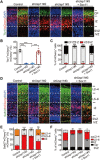
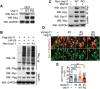
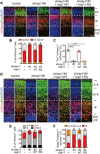
The role of protein stabilization in cortical development remains poorly understood. A recessive mutation in the USP11 gene is found in a rare neurodevelopmental disorder with intellectual disability, but its pathogenicity and molecular mechanism are unknown. Here, we show that mouse Usp11 is expressed highly in embryonic cerebral cortex, and Usp11 deficiency impairs layer 6 neuron production, delays late-born neuronal migration, and disturbs cognition and anxiety behaviors. Mechanistically, these functions are mediated by a previously unidentified Usp11 substrate, Sox11. Usp11 ablation compromises Sox11 protein accumulation in the developing cortex, despite the induction of Sox11 mRNA. The disease-associated Usp11 mutant fails to stabilize Sox11 and is unable to support cortical neurogenesis and neuronal migration. Our findings define a critical function of Usp11 in cortical development and highlight the importance of orchestrating protein stabilization mechanisms into transcription regulatory programs for a robust induction of cell fate determinants during early brain development.
Copyright © 2021 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works. Distributed under a Creative Commons Attribution NonCommercial License 4.0 (CC BY-NC).
Figures





References
-
- Florio M., Huttner W. B., Neural progenitors, neurogenesis and the evolution of the neocortex. Development 141, 2182–2194 (2014). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

