DNA Mismatch Repair and its Role in Huntington's Disease
- PMID: 33579865
- PMCID: PMC7990411
- DOI: 10.3233/JHD-200438
DNA Mismatch Repair and its Role in Huntington's Disease
Abstract
DNA mismatch repair (MMR) is a highly conserved genome stabilizing pathway that corrects DNA replication errors, limits chromosomal rearrangements, and mediates the cellular response to many types of DNA damage. Counterintuitively, MMR is also involved in the generation of mutations, as evidenced by its role in causing somatic triplet repeat expansion in Huntington's disease (HD) and other neurodegenerative disorders. In this review, we discuss the current state of mechanistic knowledge of MMR and review the roles of key enzymes in this pathway. We also present the evidence for mutagenic function of MMR in CAG repeat expansion and consider mechanistic hypotheses that have been proposed. Understanding the role of MMR in CAG expansion may shed light on potential avenues for therapeutic intervention in HD.
Keywords: DNA mismatch repair; DNA structures; Huntington’s disease; neurodegeneration; somatic expansion; triplet repeat instability.
Conflict of interest statement
The authors have no conflicts of interest.
Figures
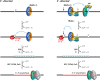

References
-
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. 2nd ed. Washington, D.C.: ASM Press; 2006.
-
- Iyer RR, Pluciennik A, Burdett V, Modrich PL. DNA mismatch repair: functions and mechanisms. Chem Rev. 2006;106(2):302–23. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

