PRMT5 inhibition disrupts splicing and stemness in glioblastoma
- PMID: 33579912
- PMCID: PMC7881162
- DOI: 10.1038/s41467-021-21204-5
PRMT5 inhibition disrupts splicing and stemness in glioblastoma
Abstract
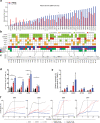
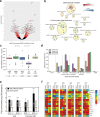
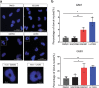
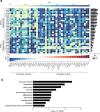
Glioblastoma (GBM) is a deadly cancer in which cancer stem cells (CSCs) sustain tumor growth and contribute to therapeutic resistance. Protein arginine methyltransferase 5 (PRMT5) has recently emerged as a promising target in GBM. Using two orthogonal-acting inhibitors of PRMT5 (GSK591 or LLY-283), we show that pharmacological inhibition of PRMT5 suppresses the growth of a cohort of 46 patient-derived GBM stem cell cultures, with the proneural subtype showing greater sensitivity. We show that PRMT5 inhibition causes widespread disruption of splicing across the transcriptome, particularly affecting cell cycle gene products. We identify a GBM splicing signature that correlates with the degree of response to PRMT5 inhibition. Importantly, we demonstrate that LLY-283 is brain-penetrant and significantly prolongs the survival of mice with orthotopic patient-derived xenografts. Collectively, our findings provide a rationale for the clinical development of brain penetrant PRMT5 inhibitors as treatment for GBM.
Conflict of interest statement
Z.Q.B. is an employee of Eli Lilly and Company. All other authors declare no competing interests.
Figures


References
-
- Stupp, R. et al. Radiotherapy plus concomitant and adjuvant temozolomide for globlastoma. N. Engl. J. Med.10.1016/j.canrad.2005.05.001 (2005). - PubMed
-
- Singh, S. K. et al. Identification of a cancer stem cell in human brain tumors. Cancer Res.10.1038/nature03128 (2003). - PubMed
-
- Bao, S. et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature10.1038/nature05236 (2006). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

