Insertion-sequence-mediated mutations both promote and constrain evolvability during a long-term experiment with bacteria
- PMID: 33579917
- PMCID: PMC7881107
- DOI: 10.1038/s41467-021-21210-7
Insertion-sequence-mediated mutations both promote and constrain evolvability during a long-term experiment with bacteria
Abstract
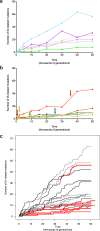
Insertion sequences (IS) are ubiquitous bacterial mobile genetic elements, and the mutations they cause can be deleterious, neutral, or beneficial. The long-term dynamics of IS elements and their effects on bacteria are poorly understood, including whether they are primarily genomic parasites or important drivers of adaptation by natural selection. Here, we investigate the dynamics of IS elements and their contribution to genomic evolution and fitness during a long-term experiment with Escherichia coli. IS elements account for ~35% of the mutations that reached high frequency through 50,000 generations in those populations that retained the ancestral point-mutation rate. In mutator populations, IS-mediated mutations are only half as frequent in absolute numbers. In one population, an exceptionally high ~8-fold increase in IS150 copy number is associated with the beneficial effects of early insertion mutations; however, this expansion later slowed down owing to reduced IS150 activity. This population also achieves the lowest fitness, suggesting that some avenues for further adaptation are precluded by the IS150-mediated mutations. More generally, across all populations, we find that higher IS activity becomes detrimental to adaptation over evolutionary time. Therefore, IS-mediated mutations can both promote and constrain evolvability.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Wagner, A. Robustness and Evolvability in Living Systems (Princeton University Press, Princeton, 2005).
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources

