Tumor cell plasticity, heterogeneity, and resistance in crucial microenvironmental niches in glioma
- PMID: 33579922
- PMCID: PMC7881116
- DOI: 10.1038/s41467-021-21117-3
Tumor cell plasticity, heterogeneity, and resistance in crucial microenvironmental niches in glioma
Abstract
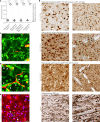
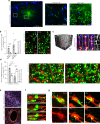
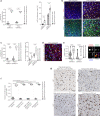
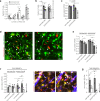
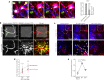
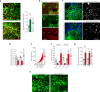
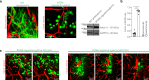
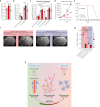
Both the perivascular niche (PVN) and the integration into multicellular networks by tumor microtubes (TMs) have been associated with progression and resistance to therapies in glioblastoma, but their specific contribution remained unknown. By long-term tracking of tumor cell fate and dynamics in the live mouse brain, differential therapeutic responses in both niches are determined. Both the PVN, a preferential location of long-term quiescent glioma cells, and network integration facilitate resistance against cytotoxic effects of radiotherapy and chemotherapy-independently of each other, but with additive effects. Perivascular glioblastoma cells are particularly able to actively repair damage to tumor regions. Population of the PVN and resistance in it depend on proficient NOTCH1 expression. In turn, NOTCH1 downregulation induces resistant multicellular networks by TM extension. Our findings identify NOTCH1 as a central switch between the PVN and network niche in glioma, and demonstrate robust cross-compensation when only one niche is targeted.
Conflict of interest statement
E.J., M.O., W.W. and F.W. report the patent (WO2017020982A1) “Agents for use in the treatment of glioma”. F.W. is co-founder of DC Europa Ltd (a company trading under the name Divide & Conquer) that is developing new medicines for the treatment of glioma. Divide & Conquer also provides research funding to F.W.’s lab under a research collaboration agreement. All other authors declare no competing interests.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

