The acquisition of molecular drivers in pediatric therapy-related myeloid neoplasms
- PMID: 33579957
- PMCID: PMC7880998
- DOI: 10.1038/s41467-021-21255-8
The acquisition of molecular drivers in pediatric therapy-related myeloid neoplasms
Abstract
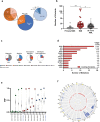
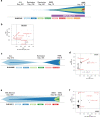
Pediatric therapy-related myeloid neoplasms (tMN) occur in children after exposure to cytotoxic therapy and have a dismal prognosis. The somatic and germline genomic alterations that drive these myeloid neoplasms in children and how they arise have yet to be comprehensively described. We use whole exome, whole genome, and/or RNA sequencing to characterize the genomic profile of 84 pediatric tMN cases (tMDS: n = 28, tAML: n = 56). Our data show that Ras/MAPK pathway mutations, alterations in RUNX1 or TP53, and KMT2A rearrangements are frequent somatic drivers, and we identify cases with aberrant MECOM expression secondary to enhancer hijacking. Unlike adults with tMN, we find no evidence of pre-existing minor tMN clones (including those with TP53 mutations), but rather the majority of cases are unrelated clones arising as a consequence of cytotoxic therapy. These studies also uncover rare cases of lineage switch disease rather than true secondary neoplasms.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

