Ultra high-risk PFA ependymoma is characterized by loss of chromosome 6q
- PMID: 33580238
- PMCID: PMC8328032
- DOI: 10.1093/neuonc/noab034
Ultra high-risk PFA ependymoma is characterized by loss of chromosome 6q
Abstract
Background: Within PF-EPN-A, 1q gain is a marker of poor prognosis, however, it is unclear if within PF-EPN-A additional cytogenetic events exist which can refine risk stratification.
Methods: Five independent non-overlapping cohorts of PF-EPN-A were analyzed applying genome-wide methylation arrays for chromosomal and clinical variables predictive of survival.
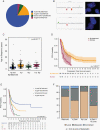
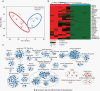
Results: Across all cohorts, 663 PF-EPN-A were identified. The most common broad copy number event was 1q gain (18.9%), followed by 6q loss (8.6%), 9p gain (6.5%), and 22q loss (6.8%). Within 1q gain tumors, there was significant enrichment for 6q loss (17.7%), 10q loss (16.9%), and 16q loss (15.3%). The 5-year progression-free survival (PFS) was strikingly worse in those patients with 6q loss, with a 5-year PFS of 50% (95% CI 45%-55%) for balanced tumors, compared with 32% (95% CI 24%-44%) for 1q gain only, 7.3% (95% CI 2.0%-27%) for 6q loss only and 0 for both 1q gain and 6q loss (P = 1.65 × 10-13). After accounting for treatment, 6q loss remained the most significant independent predictor of survival in PF-EPN-A but is not in PF-EPN-B. Distant relapses were more common in 1q gain irrespective of 6q loss. RNA sequencing comparing 6q loss to 6q balanced PF-EPN-A suggests that 6q loss forms a biologically distinct group.
Conclusions: We have identified an ultra high-risk PF-EPN-A ependymoma subgroup, which can be reliably ascertained using cytogenetic markers in routine clinical use. A change in treatment paradigm is urgently needed for this particular subset of PF-EPN-A where novel therapies should be prioritized for upfront therapy.
Keywords: 1q gain; 6q loss; PFA; PFB; ependymoma; posterior fossa; subgrouping.
© The Author(s) 2021. Published by Oxford University Press on behalf of the Society for Neuro-Oncology. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures

References
-
- Khatua S, Ramaswamy V, Bouffet E. Current therapy and the evolving molecular landscape of paediatric ependymoma. Eur J Cancer. 2017;70:34–41. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

