ARF2 represses expression of plant GRF transcription factors in a complementary mechanism to microRNA miR396
- PMID: 33580700
- PMCID: PMC8133599
- DOI: 10.1093/plphys/kiab014
ARF2 represses expression of plant GRF transcription factors in a complementary mechanism to microRNA miR396
Abstract
Members of the GROWTH REGULATING FACTOR (GRF) family of transcription factors play key roles in the promotion of plant growth and development. Many GRFs are post-transcriptionally repressed by microRNA (miRNA) miR396, an evolutionarily conserved small RNA, which restricts their expression to proliferative tissue. We performed a comprehensive analysis of the GRF family in eudicot plants and found that in many species all the GRFs have a miR396-binding site. Yet, we also identified GRFs with mutations in the sequence recognized by miR396, suggesting a partial or complete release of their post-transcriptional repression. Interestingly, Brassicaceae species share a group of GRFs that lack miR396 regulation, including Arabidopsis GRF5 and GRF6. We show that instead of miR396-mediated post-transcriptional regulation, the spatiotemporal control of GRF5 is achieved through evolutionarily conserved promoter sequences, and that AUXIN RESPONSE FACTOR 2 (ARF2) binds to such conserved sequences to repress GRF5 expression. Furthermore, we demonstrate that the unchecked expression of GRF5 in arf2 mutants is responsible for the increased cell number of arf2 leaves. The results describe a switch in the repression mechanisms that control the expression of GRFs and mechanistically link the control of leaf growth by miR396, GRFs, and ARF2 transcription factors.
© American Society of Plant Biologists 2021. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures


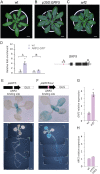

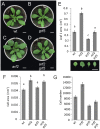
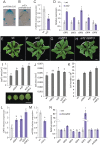

References
-
- Allen E, Xie Z, Gustafson AM, Carrington JC (2005) microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221 - PubMed
-
- Che R, Tong H, Shi B, Liu Y, Fang S, Liu D, Xiao Y, Hu B, Liu L, Wang H, et al. (2015) Control of grain size and rice yield by GL2-mediated brassinosteroid responses. Nat Plants 2: 15195. - PubMed
-
- Chitwood DH, Sinha NR (2016) Evolutionary and environmental forces sculpting leaf development. Curr Biol 26: R297–R306 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

