Dectin-1 limits autoimmune neuroinflammation and promotes myeloid cell-astrocyte crosstalk via Card9-independent expression of Oncostatin M
- PMID: 33581044
- PMCID: PMC7956124
- DOI: 10.1016/j.immuni.2021.01.004
Dectin-1 limits autoimmune neuroinflammation and promotes myeloid cell-astrocyte crosstalk via Card9-independent expression of Oncostatin M
Abstract
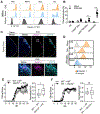
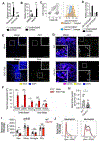
Pathologic roles of innate immunity in neurologic disorders are well described, but their beneficial aspects are less understood. Dectin-1, a C-type lectin receptor (CLR), is largely known to induce inflammation. Here, we report that Dectin-1 limited experimental autoimmune encephalomyelitis (EAE), while its downstream signaling molecule, Card9, promoted the disease. Myeloid cells mediated the pro-resolution function of Dectin-1 in EAE with enhanced gene expression of the neuroprotective molecule, Oncostatin M (Osm), through a Card9-independent pathway, mediated by the transcription factor NFAT. Furthermore, we find that the Osm receptor (OsmR) functioned specifically in astrocytes to reduce EAE severity. Notably, Dectin-1 did not respond to heat-killed Mycobacteria, an adjuvant to induce EAE. Instead, endogenous Dectin-1 ligands, including galectin-9, in the central nervous system (CNS) were involved to limit EAE. Our study reveals a mechanism of beneficial myeloid cell-astrocyte crosstalk regulated by a Dectin-1 pathway and identifies potential therapeutic targets for autoimmune neuroinflammation.
Keywords: C-type lectin receptors; CLRs; Card9; Dectin-1/Clec7a; Gal-9; Galectin-9; MS; OSMR; Oncostatin M; Osm; astrocytes; innate immunity; multiple sclerosis; neuroimmunology.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests R.-R.J. is a consultant of Boston Scientific and received a research grant from the company. This activity is not related to the current study. The remaining authors declare no competing interests.
Figures





Comment in
-
A totally OSM gift to astrocytes relieves inflammation.Immunity. 2021 Mar 9;54(3):401-403. doi: 10.1016/j.immuni.2021.02.011. Immunity. 2021. PMID: 33691129
References
-
- Bode K, Bujupi F, Link C, Hein T, Zimmermann S, Peiris D, Jaquet V, Lepenies B, Weyd H, and Krammer PH (2019). Dectin-1 Binding to Annexins on Apoptotic Cells Induces Peripheral Immune Tolerance via NADPH Oxidase-2. Cell Rep 29, 4435–4446 e4439. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

