DGCR8-dependent efficient pri-miRNA processing of human pri-miR-9-2
- PMID: 33581109
- PMCID: PMC7995608
- DOI: 10.1016/j.jbc.2021.100409
DGCR8-dependent efficient pri-miRNA processing of human pri-miR-9-2
Abstract
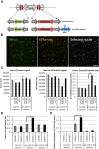
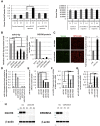
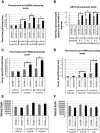
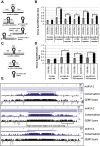
Microprocessor complex, including DiGeorge syndrome critical region gene 8 (DGCR8) and DROSHA, recognizes and cleaves primary transcripts of microRNAs (pri-miRNAs) in the maturation of canonical miRNAs. The study of DGCR8 haploinsufficiency reveals that the efficiency of this activity varies for different miRNA species. It is thought that this variation might be associated with the risk of schizophrenia with 22q11 deletion syndrome caused by disruption of the DGCR8 gene. However, the underlying mechanism for varying action of DGCR8 with each miRNA remains largely unknown. Here, we used in vivo monitoring to measure the efficiency of DGCR8-dependent microprocessor activity in cultured cells. We confirmed that this system recapitulates the microprocessor activity of endogenous pri-miRNA with expression of a ratiometric fluorescence reporter. Using this system, we detected mir-9-2 as one of the most efficient targets. We also identified a novel DGCR8-responsive RNA element, which is highly conserved among mammalian species and could be regulated at the epi-transcriptome (RNA modification) level. This unique feature between DGCR8 and pri-miR-9-2 processing may suggest a link to the risk of schizophrenia.
Keywords: DGCR8; DiGeorge syndrome; and fluorescence; miRNA; neurogenesis; pri-miR-9-2; pri-miRNA processing; ribonuclease; schizophrenia.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest H. O. is a paid member of the Scientific Advisory Board of San Bio Co, Ltd and K Pharma, Inc M. Y. is a scientific advisor of K Pharma, Inc.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

