Resource sharing between central metabolism and cell envelope synthesis
- PMID: 33581378
- PMCID: PMC7988295
- DOI: 10.1016/j.mib.2021.01.015
Resource sharing between central metabolism and cell envelope synthesis
Abstract
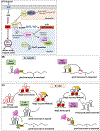
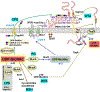
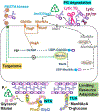
Synthesis of the bacterial cell envelope requires a regulated partitioning of resources from central metabolism. Here, we consider the key metabolic junctions that provide the precursors needed to assemble the cell envelope. Peptidoglycan synthesis requires redirection of a glycolytic intermediate, fructose-6-phosphate, into aminosugar biosynthesis by the highly regulated branchpoint enzyme GlmS. MurA directs the downstream product, UDP-GlcNAc, specifically into peptidoglycan synthesis. Other shared resources required for cell envelope synthesis include the isoprenoid carrier lipid undecaprenyl phosphate and amino acids required for peptidoglycan cross-bridges. Assembly of the envelope requires a sharing of limited resources between competing cellular pathways and may additionally benefit from scavenging of metabolites released from neighboring cells or the formation of symbiotic relationships with a host.
Copyright © 2021 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Conflict of interest statement.
Nothing declared.
Figures

References
-
- Chubukov V, Gerosa L, Kochanowski K, Sauer U: Coordination of microbial metabolism. Nat Rev Microbiol 2014, 12:327–340. - PubMed
-
- Reith J, Mayer C: Peptidoglycan turnover and recycling in Gram-positive bacteria. Appl Microbiol Biotechnol 2011, 92:1–11. - PubMed
-
-
Kawai Y, Mercier R, Mickiewicz K, Serafini A, Sorio de Carvalho LP, Errington J: Crucial role for central carbon metabolism in the bacterial L-form switch and killing by beta-lactam antibiotics. Nat Microbiol 2019, 4:1716–1726.
• Blocking PG synthesis can trigger the emergence of L-forms, bacteria that lack a cell wall. However, the resultant increase in flux of carbon into lower glycolysis can ultimately the electron transfer chain triggers oxidative stress.
-
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

