Transfer learning for establishment of recognition of COVID-19 on CT imaging using small-sized training datasets
- PMID: 33584016
- PMCID: PMC7866884
- DOI: 10.1016/j.knosys.2021.106849
Transfer learning for establishment of recognition of COVID-19 on CT imaging using small-sized training datasets
Abstract
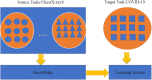
The coronavirus disease, called COVID-19, which is spreading fast worldwide since the end of 2019, and has become a global challenging pandemic. Until 27th May 2020, it caused more than 5.6 million individuals infected throughout the world and resulted in greater than 348,145 deaths. CT images-based classification technique has been tried to use the identification of COVID-19 with CT imaging by hospitals, which aims to minimize the possibility of virus transmission and alleviate the burden of clinicians and radiologists. Early diagnosis of COVID-19, which not only prevents the disease from spreading further but allows more reasonable allocation of limited medical resources. Therefore, CT images play an essential role in identifying cases of COVID-19 that are in great need of intensive clinical care. Unfortunately, the current public health emergency, which has caused great difficulties in collecting a large set of precise data for training neural networks. To tackle this challenge, our first thought is transfer learning, which is a technique that aims to transfer the knowledge from one or more source tasks to a target task when the latter has fewer training data. Since the training data is relatively limited, so a transfer learning-based DensNet-121 approach for the identification of COVID-19 is established. The proposed method is inspired by the precious work of predecessors such as CheXNet for identifying common Pneumonia, which was trained using the large Chest X-ray14 dataset, and the dataset contains 112,120 frontal chest X-rays of 14 different chest diseases (including Pneumonia) that are individually labeled and achieved good performance. Therefore, CheXNet as the pre-trained network was used for the target task (COVID-19 classification) by fine-tuning the network weights on the small-sized dataset in the target task. Finally, we evaluated our proposed method on the COVID-19-CT dataset. Experimentally, our method achieves state-of-the-art performance for the accuracy (ACC) and F1-score. The quantitative indicators show that the proposed method only uses a GPU can reach the best performance, up to 0.87 and 0.86, respectively, compared with some widely used and recent deep learning methods, which are helpful for COVID-19 diagnosis and patient triage. The codes used in this manuscript are publicly available on GitHub at (https://github.com/lichun0503/CT-Classification).
Keywords: COVID-19 Pneumonia; Classification; Small-sized samples learning; Transfer learning.
© 2021 Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




Similar articles
-
CoroNet: A deep neural network for detection and diagnosis of COVID-19 from chest x-ray images.Comput Methods Programs Biomed. 2020 Nov;196:105581. doi: 10.1016/j.cmpb.2020.105581. Epub 2020 Jun 5. Comput Methods Programs Biomed. 2020. PMID: 32534344 Free PMC article.
-
A Cascade-SEME network for COVID-19 detection in chest x-ray images.Med Phys. 2021 May;48(5):2337-2353. doi: 10.1002/mp.14711. Epub 2021 Mar 29. Med Phys. 2021. PMID: 33778966 Free PMC article.
-
TEM virus images: Benchmark dataset and deep learning classification.Comput Methods Programs Biomed. 2021 Sep;209:106318. doi: 10.1016/j.cmpb.2021.106318. Epub 2021 Jul 29. Comput Methods Programs Biomed. 2021. PMID: 34375851
-
CovXNet: A multi-dilation convolutional neural network for automatic COVID-19 and other pneumonia detection from chest X-ray images with transferable multi-receptive feature optimization.Comput Biol Med. 2020 Jul;122:103869. doi: 10.1016/j.compbiomed.2020.103869. Epub 2020 Jun 20. Comput Biol Med. 2020. PMID: 32658740 Free PMC article.
-
Development and integration of VGG and dense transfer-learning systems supported with diverse lung images for discovery of the Coronavirus identity.Inform Med Unlocked. 2022;32:101004. doi: 10.1016/j.imu.2022.101004. Epub 2022 Jul 8. Inform Med Unlocked. 2022. PMID: 35822170 Free PMC article. Review.
Cited by
-
Explainable multi-instance and multi-task learning for COVID-19 diagnosis and lesion segmentation in CT images.Knowl Based Syst. 2022 Sep 27;252:109278. doi: 10.1016/j.knosys.2022.109278. Epub 2022 Jun 27. Knowl Based Syst. 2022. PMID: 35783000 Free PMC article.
-
GLAPAL-H: Global, Local, And Parts Aware Learner for Hydrocephalus Infection Diagnosis in Low-Field MRI.medRxiv [Preprint]. 2025 Jun 6:2025.05.14.25327461. doi: 10.1101/2025.05.14.25327461. medRxiv. 2025. Update in: IEEE Trans Biomed Eng. 2025 Jun 09;PP. doi: 10.1109/TBME.2025.3578541. PMID: 40463531 Free PMC article. Updated. Preprint.
-
Multi-task semantic segmentation of CT images for COVID-19 infections using DeepLabV3+ based on dilated residual network.Phys Eng Sci Med. 2022 Jun;45(2):443-455. doi: 10.1007/s13246-022-01110-w. Epub 2022 Mar 14. Phys Eng Sci Med. 2022. PMID: 35286619 Free PMC article.
-
How much BiGAN and CycleGAN-learned hidden features are effective for COVID-19 detection from CT images? A comparative study.J Supercomput. 2023;79(3):2850-2881. doi: 10.1007/s11227-022-04775-y. Epub 2022 Aug 26. J Supercomput. 2023. PMID: 36042937 Free PMC article.
-
A COMPARATIVE STUDY OF X-RAY AND CT IMAGES IN COVID-19 DETECTION USING IMAGE PROCESSING AND DEEP LEARNING TECHNIQUES.Comput Methods Programs Biomed Update. 2022;2:100054. doi: 10.1016/j.cmpbup.2022.100054. Epub 2022 Mar 7. Comput Methods Programs Biomed Update. 2022. PMID: 35281724 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
