Genomic Characterization of Multidrug-Resistant Escherichia coli BH100 Sub-strains
- PMID: 33584554
- PMCID: PMC7874104
- DOI: 10.3389/fmicb.2020.549254
Genomic Characterization of Multidrug-Resistant Escherichia coli BH100 Sub-strains
Abstract
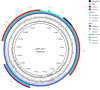
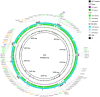
The rapid emergence of multidrug-resistant (MDR) bacteria is a global health problem. Mobile genetic elements like conjugative plasmids, transposons, and integrons are the major players in spreading resistance genes in uropathogenic Escherichia coli (UPEC) pathotype. The E. coli BH100 strain was isolated from the urinary tract of a Brazilian woman in 1974. This strain presents two plasmids carrying MDR cassettes, pBH100, and pAp, with conjugative and mobilization properties, respectively. However, its transposable elements have not been characterized. In this study, we attempted to unravel the factors involved in the mobilization of virulence and drug-resistance genes by assessing genomic rearrangements in four BH100 sub-strains (BH100 MG2014, BH100 MG2017, BH100L MG2017, and BH100N MG2017). Therefore, the complete genomes of the BH100 sub-strains were achieved through Next Generation Sequencing and submitted to comparative genomic analyses. Our data shows recombination events between the two plasmids in the sub-strain BH100 MG2017 and between pBH100 and the chromosome in BH100L MG2017. In both cases, IS3 and IS21 elements were detected upstream of Tn21 family transposons associated with MDR genes at the recombined region. These results integrated with Genomic island analysis suggest pBH100 might be involved in the spreading of drug resistance through the formation of resistance islands. Regarding pathogenicity, our results reveal that BH100 strain is closely related to UPEC strains and contains many IS3 and IS21-transposase-enriched genomic islands associated with virulence. This study concludes that those IS elements are vital for the evolution and adaptation of BH100 strain.
Keywords: UPEC; antibiotic resistance; genomic sequencing; mobile genetic elements; urinary tract infection.
Copyright © 2021 Carvalho, Aburjaile, Canario, Nascimento, Chartone-Souza, de Jesus, Zamyatnin, Brenig, Barh, Ghosh, Goes-Neto, Figueiredo, Soares, Ramos, Pinto and Azevedo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
Low distribution of integrons among multidrug resistant E. coli strains isolated from children with community-acquired urinary tract infections in Shiraz, Iran.Pol J Microbiol. 2008;57(3):193-8. Pol J Microbiol. 2008. PMID: 19004239
-
Defining genomic islands and uropathogen-specific genes in uropathogenic Escherichia coli.J Bacteriol. 2007 May;189(9):3532-46. doi: 10.1128/JB.01744-06. Epub 2007 Mar 9. J Bacteriol. 2007. PMID: 17351047 Free PMC article.
-
IncA/C Conjugative Plasmids Mobilize a New Family of Multidrug Resistance Islands in Clinical Vibrio cholerae Non-O1/Non-O139 Isolates from Haiti.mBio. 2016 Jul 19;7(4):e00509-16. doi: 10.1128/mBio.00509-16. mBio. 2016. PMID: 27435459 Free PMC article.
-
Pathogenomics of uropathogenic Escherichia coli.Indian J Med Microbiol. 2012 Apr-Jun;30(2):141-9. doi: 10.4103/0255-0857.96657. Indian J Med Microbiol. 2012. PMID: 22664427 Review.
-
The molecular basis of infectious diseases: pathogenicity islands and other mobile genetic elements. A review.Acta Microbiol Immunol Hung. 2003;50(4):321-30. doi: 10.1556/AMicr.50.2003.4.1. Acta Microbiol Immunol Hung. 2003. PMID: 14750434 Review.
Cited by
-
Impact of Heavy Metal and Resistance Genes on Antimicrobial Resistance: Ecological and Public Health Implications.Genes (Basel). 2025 May 24;16(6):625. doi: 10.3390/genes16060625. Genes (Basel). 2025. PMID: 40565518 Free PMC article. Review.
-
Characterization of virulence and antimicrobial resistance genes of Aeromonas media strain SD/21-15 from marine sediments in comparison with other Aeromonas spp.Front Microbiol. 2022 Dec 1;13:1022639. doi: 10.3389/fmicb.2022.1022639. eCollection 2022. Front Microbiol. 2022. PMID: 36532448 Free PMC article.
-
Molecular Factors and Mechanisms Driving Multidrug Resistance in Uropathogenic Escherichia coli-An Update.Genes (Basel). 2022 Aug 6;13(8):1397. doi: 10.3390/genes13081397. Genes (Basel). 2022. PMID: 36011308 Free PMC article. Review.
-
Understanding the intricacies of microbial biofilm formation and its endurance in chronic infections: a key to advancing biofilm-targeted therapeutic strategies.Arch Microbiol. 2024 Feb 1;206(2):85. doi: 10.1007/s00203-023-03802-7. Arch Microbiol. 2024. PMID: 38300317 Review.
-
Uropathogenic Escherichia coli (UPEC)-Associated Urinary Tract Infections: The Molecular Basis for Challenges to Effective Treatment.Microorganisms. 2023 Aug 28;11(9):2169. doi: 10.3390/microorganisms11092169. Microorganisms. 2023. PMID: 37764013 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

