Cold Adaptation Mechanisms of a Snow Alga Chlamydomonas nivalis During Temperature Fluctuations
- PMID: 33584575
- PMCID: PMC7874021
- DOI: 10.3389/fmicb.2020.611080
Cold Adaptation Mechanisms of a Snow Alga Chlamydomonas nivalis During Temperature Fluctuations
Abstract
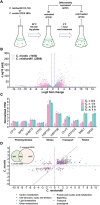
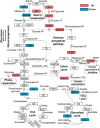
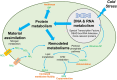
Cold environments, such as glaciers and alpine regions, constitute unique habitats for organisms living on Earth. In these harsh ecosystems, snow algae survive, florish, and even become primary producers for microbial communities. How the snow algae maintain physiological activity during violent ambient temperature changes remains unsolved. To explore the cold adaptation mechanisms of the unicellular snow alga Chlamydomonas nivalis, we compared its physiological responses to a model organism from the same genus, Chlamydomonas reinhardtii. When both cell types were exposed to a shift from 22°C to 4°C, C. nivalis exhibited an apparent advantage in cold tolerance over C. reinhardtii, as C. nivalis had both a higher growth rate and photosynthetic efficiency. To determine the cold tolerance mechanisms of C. nivalis, RNA sequencing was used to compare transcriptomes of both species after 1 h of cold treatment, mimicking temperature fluctuations in the polar region. Differential expression analysis showed that C. nivalis had fewer transcriptomic changes and was more stable during rapid temperature decrease relative to C. reinhardtii, especially for the expression of photosynthesis related genes. Additionally, we found that transcription in C. nivalis was precisely regulated by the cold response network, consisting of at least 12 transcription factors and 3 RNA-binding proteins. Moreover, genes participating in nitrogen metabolism, the pentose phosphate pathway, and polysaccharide biosynthesis were upregulated, indicating that increasing resource assimilation and remodeling of metabolisms were critical for cold adaptation in C. nivalis. Furthermore, we identified horizontally transferred genes differentially expressed in C. nivalis, which are critical for cold adaptation in other psychrophiles. Our results reveal that C. nivalis adapts rapid temperature decrease by efficiently regulating transcription of specific genes to optimize resource assimilation and metabolic pathways, providing critical insights into how snow algae survive and propagate in cold environments.
Keywords: Chlamydomonas reinhardtii; RNA-seq; cold adaptation; horizontal gene transfer; snow algae.
Copyright © 2021 Peng, Liu and Huang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
-
- Alexa A., Rahnenfuhrer J. (2010). topGO: Enrichment Analysis for Gene Ontology. R package version 2(0), 2010.
LinkOut - more resources
Full Text Sources
Other Literature Sources

