Whole-Genome Phylogenetic Analysis Reveals a Wide Diversity of Non-O157 STEC Isolated From Ground Beef and Cattle Feces
- PMID: 33584592
- PMCID: PMC7874142
- DOI: 10.3389/fmicb.2020.622663
Whole-Genome Phylogenetic Analysis Reveals a Wide Diversity of Non-O157 STEC Isolated From Ground Beef and Cattle Feces
Abstract
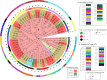
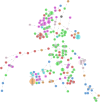
Shiga toxin-producing Escherichia coli (STEC) causes foodborne outbreaks that can lead to complications such as hemolytic uremic syndrome. Their main reservoir is cattle, and ground beef has been frequently associated with disease and outbreaks. In this study, we attempted to understand the genetic relationship among STEC isolated in Chile from different sources, their relationship to STEC from the rest of the world, and to identify molecular markers of Chilean STEC. We sequenced 62 STEC isolated in Chile using MiSeq Illumina. In silico typing was determined using tools of the Center Genomic Epidemiology, Denmark University (CGE/DTU). Genomes of our local STEC collection were compared with 113 STEC isolated worldwide through a core genome MLST (cgMLST) approach, and we also searched for distinct genes to be used as molecular markers of Chilean isolates. Genomes in our local collection were grouped based on serogroup and sequence type, and clusters were formed within local STEC. In the worldwide STEC analysis, Chilean STEC did not cluster with genomes of the rest of the world suggesting that they are not phylogenetically related to previously described STEC. The pangenome of our STEC collection was 11,650 genes, but we did not identify distinct molecular markers of local STEC. Our results showed that there may be local emerging STEC with unique features, nevertheless, no molecular markers were detected. Therefore, there might be elements such as a syntenic organization that might explain differential clustering detected between local and worldwide STEC.
Keywords: STEC; WGS; diversity; genomics; non-O157 E. coli.
Copyright © 2021 Gutiérrez, Díaz, Reyes-Jara, Yang, Meng, González-Escalona and Toro.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Álvarez-Suárez M.-E., Otero A., García-López M.-L., Dahbi G., Blanco M., Mora A., et al. (2016). Genetic characterization of Shiga toxin-producing Escherichia coli (STEC) and atypical enteropathogenic Escherichia coli (EPEC) isolates from goat’s milk and goat farm environment. Int. J. Food Microbiol. 236 148–154. 10.1016/j.ijfoodmicro.2016.07.035 - DOI - PubMed
-
- Bando S. Y., Iamashita P., Guth B. E., Dos Santos L. F., Fujita A., Abe C. M., et al. (2017). A hemolytic-uremic syndrome-associated strain O113:H21 Shiga toxin-producing Escherichia coli specifically expresses a transcriptional module containing dicA and is related to gene network dysregulation in Caco-2 cells. PLoS One 12:e0189613. 10.1371/journal.pone.0189613 - DOI - PMC - PubMed
-
- Blanco M., Padola N. L., Krüger A., Sanz M. E., Blanco J. E., González E. A., et al. (2004). Virulence genes and intimin types of Shiga-toxin-producing Escherichia coli isolated from cattle and beef products in Argentina. Int. Microbiol. Off. J. Spanish Soc. Microbiol. 7 269–276. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

