An Apical Meristem-Targeted in planta Transformation Method for the Development of Transgenics in Flax (Linum usitatissimum): Optimization and Validation
- PMID: 33584740
- PMCID: PMC7876084
- DOI: 10.3389/fpls.2020.562056
An Apical Meristem-Targeted in planta Transformation Method for the Development of Transgenics in Flax (Linum usitatissimum): Optimization and Validation
Abstract
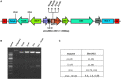
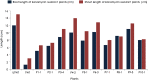
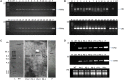
Efficient regeneration of explants devoid of intrinsic somaclonal variations is a cardinal step in plant tissue culture, thus, a vital component of transgenic technology. However, recalcitrance of economically important crops to tissue culture-based organogenesis ensues a setback in the use of transgenesis in the genetic engineering of crop plants. The present study developed an optimized, genotype-independent, nonconventional tissue culture-independent in planta strategy for the genetic transformation of flax/linseed. This apical meristem-targeted in planta transformation protocol will accelerate value addition in the dual purpose industrially important but recalcitrant fiber crop flax/linseed. The study delineated optimization of Agrobacterium tumefaciens-mediated transformation and stable T-DNA (pCambia2301:GUS:nptII) integration in flax. It established successful use of a stringent soilrite-based screening in the presence of 30 mg/L kanamycin for the identification of putative transformants. The amenability, authenticity, and reproducibility of soilrite-based kanamycin screening were further verified at the molecular level by GUS histochemical analysis of T0 seedlings, GUS and nptII gene-specific PCR, genomic Southern hybridization for stable integration of T-DNA, and expression analysis of transgenes by sqRT-PCR. This method resulted in a screening efficiency of 6.05% in the presence of kanamycin, indicating amenability of in planta flax transformation. The strategy can be a promising tool for the successful development of transgenics in flax.
Keywords: GM crops; GUS; apical meristem; in planta transformation; nptII; transgenic flax/linseed.
Copyright © 2021 Kesiraju, Tyagi, Mukherjee, Rai, Singh, Sreevathsa and Dash.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Amenability of an Agrobacterium tumefaciens-mediated shoot apical meristem-targeted in planta transformation strategy in Mango (Mangifera indica L.).GM Crops Food. 2022 Dec 31;13(1):342-354. doi: 10.1080/21645698.2022.2141014. GM Crops Food. 2022. PMID: 36421001 Free PMC article.
-
Agrobacterium tumefaciens-mediated in planta transformation strategy for development of transgenics in cotton (Gossypium hirsutum L.) with GFP as a visual marker.Physiol Mol Biol Plants. 2020 Nov;26(11):2319-2327. doi: 10.1007/s12298-020-00887-y. Epub 2020 Oct 19. Physiol Mol Biol Plants. 2020. PMID: 33268932 Free PMC article.
-
Agrobacterium tumefaciens-mediated transformation of eggplant (Solanum melongena L.) using root explants.Plant Cell Rep. 2003 Feb;21(6):549-54. doi: 10.1007/s00299-002-0546-9. Epub 2003 Jan 8. Plant Cell Rep. 2003. PMID: 12789429
-
Flax (Linum usitatissimum L.): A Potential Candidate for Phytoremediation? Biological and Economical Points of View.Plants (Basel). 2020 Apr 13;9(4):496. doi: 10.3390/plants9040496. Plants (Basel). 2020. PMID: 32294947 Free PMC article. Review.
-
Agrobacterium-mediated gene transfer in plants and biosafety considerations.Appl Biochem Biotechnol. 2012 Dec;168(7):1953-75. doi: 10.1007/s12010-012-9910-6. Epub 2012 Oct 23. Appl Biochem Biotechnol. 2012. PMID: 23090683 Review.
Cited by
-
Generation of High-Value Genomic Resource in Rice: A "Subgenomic Library" of Low-Light Tolerant Rice Cultivar Swarnaprabha.Biology (Basel). 2023 Mar 10;12(3):428. doi: 10.3390/biology12030428. Biology (Basel). 2023. PMID: 36979120 Free PMC article.
-
GWAS supported by computer vision identifies large numbers of candidate regulators of in planta regeneration in Populus trichocarpa.G3 (Bethesda). 2024 Apr 3;14(4):jkae026. doi: 10.1093/g3journal/jkae026. G3 (Bethesda). 2024. PMID: 38325329 Free PMC article.
-
A fast and genotype-independent in planta Agrobacterium-mediated transformation method for soybean.Plant Commun. 2024 Dec 9;5(12):101063. doi: 10.1016/j.xplc.2024.101063. Epub 2024 Aug 13. Plant Commun. 2024. PMID: 39138866 Free PMC article.
-
Construction and optimization of a genetic transformation system for efficient expression of human insulin-GFP fusion gene in flax.Bioresour Bioprocess. 2024 Aug 27;11(1):83. doi: 10.1186/s40643-024-00799-9. Bioresour Bioprocess. 2024. PMID: 39190215 Free PMC article.
-
Development of high-throughput tissue culture-free plant transformation systems.Plant J. 2025 Jan;121(1):e17163. doi: 10.1111/tpj.17163. Epub 2024 Dec 9. Plant J. 2025. PMID: 39652509 Free PMC article. Review.
References
-
- Aycan M., Beyaz R., Bahadir A., Yildiz M. (2019). The effect of magnetic field strength on shoot regeneration and Agrobacterium tumefaciens-mediated gene transfer in flax (Linum usitatissimum L.). Czech. J. Genet. Plant Breed. 55 20–27. 10.17221/195/2017-cjgpb - DOI
-
- Bastaki N., Cullis C. A. (2019). “Flax transformation via floral-dipping,” in Genetics and Genomics of Linum, ed. Christopher A. C. (Switzerland: Springer Nature; ), 195–214. 10.1007/978-3-030-23964-0_12 - DOI
-
- Beranova M., Rakousky S., Vavrova Z., Skalicky T. (2008). Sonication assisted Agrobacterium-mediated transformation enhances the transformation efficiency in flax (Linum usitatissimum L.). Plant Cell Tissue Organ Cult. 94 253–259. 10.1007/s11240-007-9335-z - DOI
-
- Beyaz R., Aycan M., Kayan M., Yildiz M. (2016a). A novel method for high-frequency transgenic shoot regeneration via Agrobacterium tumefaciens in flax (Linum usitatissimum L.). J. Plant Biotechnol. 43 240–247. 10.5010/jpb.2016.43.2.240 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

