Tapping Diversity From the Wild: From Sampling to Implementation
- PMID: 33584776
- PMCID: PMC7873362
- DOI: 10.3389/fpls.2021.626565
Tapping Diversity From the Wild: From Sampling to Implementation
Abstract
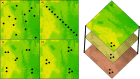
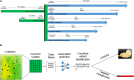
The diversity observed among crop wild relatives (CWRs) and their ability to flourish in unfavorable and harsh environments have drawn the attention of plant scientists and breeders for many decades. However, it is also recognized that the benefit gained from using CWRs in breeding is a potential rose between thorns of detrimental genetic variation that is linked to the trait of interest. Despite the increased interest in CWRs, little attention was given so far to the statistical, analytical, and technical considerations that should guide the sampling design, the germplasm characterization, and later its implementation in breeding. Here, we review the entire process of sampling and identifying beneficial genetic variation in CWRs and the challenge of using it in breeding. The ability to detect beneficial genetic variation in CWRs is strongly affected by the sampling design which should be adjusted to the spatial and temporal variation of the target species, the trait of interest, and the analytical approach used. Moreover, linkage disequilibrium is a key factor that constrains the resolution of searching for beneficial alleles along the genome, and later, the ability to deplete linked deleterious genetic variation as a consequence of genetic drag. We also discuss how technological advances in genomics, phenomics, biotechnology, and data science can improve the ability to identify beneficial genetic variation in CWRs and to exploit it in strive for higher-yielding and sustainable crops.
Keywords: breeding; crop wild relative; genetic drag; introgression; sampling design.
Copyright © 2021 Hübner and Kantar.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Aljane F., Essid A., Nahdi S. (2018). “Improvement of Fig (Ficus carica L.) by Conventional Breeding and Biotechnology,” in Advances in Plant Breeding Strategies: Fruits, eds Jain S. M., Al-Khayri J. M., Johnson D. V. (Cham: Springer; ), 343–375. 10.1007/978-3-319-91944-7_9 - DOI
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

