Profiling of MicroRNAs and Their Targets in Roots and Shoots Reveals a Potential MiRNA-Mediated Interaction Network in Response to Phosphate Deficiency in the Forestry Tree Betula luminifera
- PMID: 33584823
- PMCID: PMC7876418
- DOI: 10.3389/fgene.2021.552454
Profiling of MicroRNAs and Their Targets in Roots and Shoots Reveals a Potential MiRNA-Mediated Interaction Network in Response to Phosphate Deficiency in the Forestry Tree Betula luminifera
Abstract
Inorganic phosphate (Pi) is often lacking in natural and agro-climatic environments, which impedes the growth of economically important woody species. Plants have developed strategies to cope with low Pi (LP) availability. MicroRNAs (miRNAs) play important roles in responses to abiotic stresses, including nutrition stress, by regulating target gene expression. However, the miRNA-mediated regulation of these adaptive responses and their underlying coordinating signals are still poorly understood in forestry trees such as Betula luminifera. Transcriptomic libraries, small RNA (sRNA) libraries, and a mixed degradome cDNA library of B. luminifera roots and shoots treated under LP and normal conditions (CK) were constructed and sequenced using next-generation deep sequencing. A comprehensive B. luminifera transcriptome derived from its roots and shoots was constructed, and a total of 76,899 unigenes were generated. Analysis of the transcriptome identified 8,095 and 5,584 differentially expressed genes in roots and shoots, respectively, under LP conditions. sRNA sequencing analyses indicated that 66 and 60 miRNAs were differentially expressed in roots and shoots, respectively, under LP conditions. A total of 109 and 112 miRNA-target pairs were further validated in the roots and shoots, respectively, using degradome sequencing. Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis of differential miRNA targets indicated that the "ascorbate and aldarate metabolism" pathway responded to LP, suggesting miRNA-target pairs might participating in the removing of reactive oxidative species under LP stress. Moreover, a putative network of miRNA-target interactions involved in responses to LP stress in B. luminifera is proposed. Taken together, these findings provide useful information to decipher miRNA functions and establish a framework for exploring P signaling networks regulated by miRNAs in B. luminifera and other woody plants. It may provide new insights into the genetic engineering of high use efficiency of Pi in forestry trees.
Keywords: Betula luminifera; Pi deficiency; abiotic stress; degradome; miRNA; transcriptome.
Copyright © 2021 Zhang, Lin, Wu, Zhang, Cheng, Huang and Tong.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
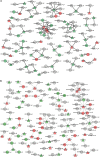
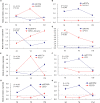
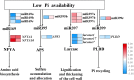
Figures

References
-
- Bai Q. Q., Wang X. Y., Chen X., Shi G. Q., Liu Z. P., Guo C. J., et al. (2018). Wheat miRNA TaemiR408 acts as an essential mediator in plant tolerance to Pi deprivation and salt stress via modulating stress-associated physiological processes. Front. Plant Sci. 9:499. 10.3389/fpls.2018.00499 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

