Inducing Non-genetically Modified Induced Embryonic Sertoli Cells Derived From Embryonic Stem Cells With Recombinant Protein Factors
- PMID: 33585437
- PMCID: PMC7875124
- DOI: 10.3389/fcell.2020.533543
Inducing Non-genetically Modified Induced Embryonic Sertoli Cells Derived From Embryonic Stem Cells With Recombinant Protein Factors
Abstract
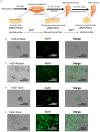
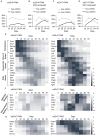
Embryonic Sertoli cells (eSCs) possess multiple supporting functions and research value in gonadal development and sex determination. However, the limitation of acquiring quality eSCs had hindered the further application. Herein, we successfully derived non-genetically modified (non-GM)-induced embryonic Sertoli-like cells (eSLCs) from mouse embryonic stem cells (ESCs) with a TM4 cell-derived conditioned medium containing recombinant endogenous protein factors Sry, Sox9, Sf1, Wt1, Gata4, and Dmrt1. These eSLCs were determined through morphology; transcriptional expression levels of stage-specific, epithelial, and mesenchymal marker genes; flow cytometry, immunofluorescence; and immunocytochemistry and functionally determined by coculture with spermatogonia stem cells. Results indicated that these eSLCs performed similarly to eSCs in specific biomarkers and expression of marker genes and supported the maturation of spermatogonia. The study induced eSLCs from mouse ESCs by defined protein factors. However, the inducing efficiency of the non-GM method was still lower than that of the lentiviral transduction method. Thus, this work established a foundation for future production of non-GM eSLCs for clinical applications and fundamental theory research.
Keywords: Sertoli cells; embryonic stem cells; lentiviral transduction; non-genetically modified; recombinant endogenous protein factors.
Copyright © 2021 Xu, Mohsin, Luo, Xie, Peng, Wang, Ahmed, Hang, Zhuang and Guo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

