Specific and Rapid SARS-CoV-2 Identification Based on LC-MS/MS Analysis
- PMID: 33585737
- PMCID: PMC7857140
- DOI: 10.1021/acsomega.0c04691
Specific and Rapid SARS-CoV-2 Identification Based on LC-MS/MS Analysis
Abstract
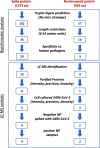
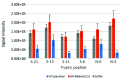
SARS-CoV-2, the etiologic agent of the COVID-19 pandemic, emerged as the cause of a global crisis. Rapid and reliable clinical diagnosis is essential for effectively controlling transmission. The gold standard assay for SARS-CoV-2 identification is the highly sensitive real-time quantitative polymerase chain reaction (RT-qPCR); however, this assay depends on specialized reagents and may suffer from false results. Thus, additional assays based on different approaches could be beneficial. Here, we present a novel method for SARS-CoV-2 identification based on mass spectrometry. The approach we implemented combines a multistep procedure for the rational down-selection of a set of reliable markers out of all optional in silico derived tryptic peptides in viral proteins, followed by monitoring of peptides derived from tryptic digests of purified proteins, cell-cultured SARS-CoV-2, and nasopharyngeal (NP) swab matrix spiked with the virus. The marker selection was based on specificity to SARS-CoV-2 and on analytical parameters including sensitivity, linearity, and reproducibility. The final assay is based on six unique and specific peptide markers for SARS-CoV-2 identification. The simple and rapid (2.5 h) protocol we developed consists of virus heat inactivation and denaturation, tryptic digestion, and identification of the selected markers by liquid chromatography coupled to high-resolution mass spectrometry (LC-MS/MS). The developed assay enabled the identification of 104 PFU/mL SARS-CoV-2 spiked into buffer. Finally, the assay was successfully applied to 16 clinical samples diagnosed by RT-qPCR, achieving 94% concordance with the current gold standard assay. To conclude, the novel MS-based assay described here is specific, rapid, simple, and is believed to provide a complementary assay to the RT-qPCR method.
© 2021 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


Similar articles
-
Coupling immuno-magnetic capture with LC-MS/MS(MRM) as a sensitive, reliable, and specific assay for SARS-CoV-2 identification from clinical samples.Anal Bioanal Chem. 2022 Feb;414(5):1949-1962. doi: 10.1007/s00216-021-03831-5. Epub 2022 Jan 4. Anal Bioanal Chem. 2022. PMID: 34981149 Free PMC article.
-
Quantitative Assessment of SARS-CoV-2 Virus in Nasopharyngeal Swabs Stored in Transport Medium by a Straightforward LC-MS/MS Assay Targeting Nucleocapsid, Membrane, and Spike Proteins.J Proteome Res. 2021 Feb 5;20(2):1434-1443. doi: 10.1021/acs.jproteome.0c00887. Epub 2021 Jan 26. J Proteome Res. 2021. PMID: 33497234
-
Operation Moonshot: rapid translation of a SARS-CoV-2 targeted peptide immunoaffinity liquid chromatography-tandem mass spectrometry test from research into routine clinical use.Clin Chem Lab Med. 2022 Nov 17;61(2):302-310. doi: 10.1515/cclm-2022-1000. Print 2023 Jan 27. Clin Chem Lab Med. 2022. PMID: 36395058
-
Identification of SARS-CoV-2 Proteins from Nasopharyngeal Swabs Probed by Multiple Reaction Monitoring Tandem Mass Spectrometry.ACS Omega. 2021 Dec 7;6(50):34945-34953. doi: 10.1021/acsomega.1c05587. eCollection 2021 Dec 21. ACS Omega. 2021. PMID: 34926968 Free PMC article.
-
Advances in rapid detection of SARS-CoV-2 by mass spectrometry.Trends Analyt Chem. 2022 Dec;157:116759. doi: 10.1016/j.trac.2022.116759. Epub 2022 Aug 20. Trends Analyt Chem. 2022. PMID: 36035092 Free PMC article. Review.
Cited by
-
Evaluation of Variant-Specific Peptides for Detection of SARS-CoV-2 Variants of Concern.J Proteome Res. 2022 Oct 7;21(10):2443-2452. doi: 10.1021/acs.jproteome.2c00325. Epub 2022 Sep 15. J Proteome Res. 2022. PMID: 36108102 Free PMC article.
-
Modified ELISA for Ultrasensitive Diagnosis.J Clin Med. 2021 Nov 7;10(21):5197. doi: 10.3390/jcm10215197. J Clin Med. 2021. PMID: 34768717 Free PMC article. Review.
-
Targeted Detection of SARS-CoV-2 Nucleocapsid Sequence Variants by Mass Spectrometric Analysis of Tryptic Peptides.J Proteome Res. 2022 Jan 7;21(1):142-150. doi: 10.1021/acs.jproteome.1c00613. Epub 2021 Nov 15. J Proteome Res. 2022. PMID: 34779632 Free PMC article.
-
Development of an immunofluorescence assay for detection of SARS-CoV-2.Arch Virol. 2022 Apr;167(4):1041-1049. doi: 10.1007/s00705-022-05392-z. Epub 2022 Feb 22. Arch Virol. 2022. PMID: 35192015 Free PMC article.
-
Discriminative Identification of SARS-CoV-2 Variants Based on Mass-Spectrometry Analysis.Biomedicines. 2023 Aug 24;11(9):2373. doi: 10.3390/biomedicines11092373. Biomedicines. 2023. PMID: 37760814 Free PMC article.
References
-
- Wu F.; Zhao S.; Yu B.; Chen Y. M.; Wang W.; Song Z. G.; Hu Y.; Tao Z. W.; Tian J. H.; Pei Y. Y.; Yuan M. L.; Zhang Y. L.; Dai F. H.; Liu Y.; Wang Q. M.; Zheng J. J.; Xu L.; Holmes E. C.; Zhang Y. Z. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. 10.1038/s41586-020-2008-3. - DOI - PMC - PubMed
-
- Zhou P.; Yang X. L.; Wang X. G.; Hu B.; Zhang L.; Zhang W.; Si H. R.; Zhu Y.; Li B.; Huang C. L.; Chen H. D.; Chen J.; Luo Y.; Guo H.; Jiang R. D.; Liu M. Q.; Chen Y.; Shen X. R.; Wang X.; Zheng X. S.; Zhao K.; Chen Q. J.; Deng F.; Liu L. L.; Yan B.; Zhan F. X.; Wang Y. Y.; Xiao G. F.; Shi Z. L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. 10.1038/s41586-020-2012-7. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

