Comparative genomics of white and opaque cell states supports an epigenetic mechanism of phenotypic switching in Candida albicans
- PMID: 33585874
- PMCID: PMC8366294
- DOI: 10.1093/g3journal/jkab001
Comparative genomics of white and opaque cell states supports an epigenetic mechanism of phenotypic switching in Candida albicans
Abstract
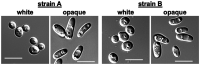
Several Candida species can undergo a heritable and reversible transition from a 'white' state to a mating proficient 'opaque' state. This ability relies on highly interconnected transcriptional networks that control cell-type-specific gene expression programs over multiple generations. Candida albicans, the most prominent pathogenic Candida species, provides a well-studied paradigm for the white-opaque transition. In this species, a network of at least eight transcriptional regulators controls the balance between white and opaque states that have distinct morphologies, transcriptional profiles, and physiological properties. Given the reversible nature and the high frequency of white-opaque transitions, it is widely assumed that this switch is governed by epigenetic mechanisms that occur independently of any changes in DNA sequence. However, a direct genomic comparison between white and opaque cells has yet to be performed. Here, we present a whole-genome comparative analysis of C. albicans white and opaque cells. This analysis revealed rare genetic changes between cell states, none of which are linked to white-opaque switching. This result is consistent with epigenetic mechanisms controlling cell state differentiation in C. albicans and provides direct evidence against a role for genetic variation in mediating the switch.
Keywords: Candida albicans; comparative genomics; epigenetic switch.
© The Author(s) 2021. Published by Oxford University Press on behalf of Genetics Society of America.
Figures


Similar articles
-
Ssn6 Defines a New Level of Regulation of White-Opaque Switching in Candida albicans and Is Required For the Stochasticity of the Switch.mBio. 2016 Jan 26;7(1):e01565-15. doi: 10.1128/mBio.01565-15. mBio. 2016. PMID: 26814177 Free PMC article.
-
Structure of the transcriptional network controlling white-opaque switching in Candida albicans.Mol Microbiol. 2013 Oct;90(1):22-35. doi: 10.1111/mmi.12329. Epub 2013 Aug 25. Mol Microbiol. 2013. PMID: 23855748 Free PMC article.
-
Temporal anatomy of an epigenetic switch in cell programming: the white-opaque transition of C. albicans.Mol Microbiol. 2010 Oct;78(2):331-43. doi: 10.1111/j.1365-2958.2010.07331.x. Epub 2010 Aug 29. Mol Microbiol. 2010. PMID: 20735781 Free PMC article.
-
Regulation of white-opaque switching in Candida albicans.Med Microbiol Immunol. 2010 Aug;199(3):165-72. doi: 10.1007/s00430-010-0147-0. Med Microbiol Immunol. 2010. PMID: 20390300 Review.
-
White-opaque switching in Candida albicans: cell biology, regulation, and function.Microbiol Mol Biol Rev. 2024 Jun 27;88(2):e0004322. doi: 10.1128/mmbr.00043-22. Epub 2024 Mar 28. Microbiol Mol Biol Rev. 2024. PMID: 38546228 Free PMC article. Review.
Cited by
-
Variation in transcription regulator expression underlies differences in white-opaque switching between the SC5314 reference strain and the majority of Candida albicans clinical isolates.Genetics. 2023 Nov 1;225(3):iyad162. doi: 10.1093/genetics/iyad162. Genetics. 2023. PMID: 37811798 Free PMC article.
-
Cell-type memory in a single-cell eukaryote requires the continuous presence of a specific transcription regulator.Proc Natl Acad Sci U S A. 2023 May 23;120(21):e2220568120. doi: 10.1073/pnas.2220568120. Epub 2023 May 15. Proc Natl Acad Sci U S A. 2023. PMID: 37186823 Free PMC article.
-
Farnesol and phosphorylation of the transcriptional regulator Efg1 affect Candida albicans white-opaque switching rates.PLoS One. 2023 Jan 20;18(1):e0280233. doi: 10.1371/journal.pone.0280233. eCollection 2023. PLoS One. 2023. PMID: 36662710 Free PMC article.
-
Strain background of Candida albicans interacts with SIR2 to alter phenotypic switching.Microbiology (Reading). 2024 Mar;170(3):001444. doi: 10.1099/mic.0.001444. Microbiology (Reading). 2024. PMID: 38446018 Free PMC article.
-
Showcasing Fungal Genetics & Genomics with the Genetics Society of America.Genetics. 2021 Feb 9;217(2):iyaa034. doi: 10.1093/genetics/iyaa034. Genetics. 2021. PMID: 33724422 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
