COVID-19: immunopathology, pathophysiological mechanisms, and treatment options
- PMID: 33586189
- PMCID: PMC8013908
- DOI: 10.1002/path.5642
COVID-19: immunopathology, pathophysiological mechanisms, and treatment options
Abstract
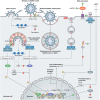
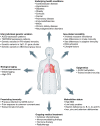
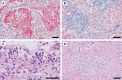
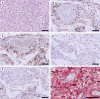
Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), continues to spread globally despite the worldwide implementation of preventive measures to combat the disease. Although most COVID-19 cases are characterised by a mild, self-limiting disease course, a considerable subset of patients develop a more severe condition, varying from pneumonia and acute respiratory distress syndrome (ARDS) to multi-organ failure (MOF). Progression of COVID-19 is thought to occur as a result of a complex interplay between multiple pathophysiological mechanisms, all of which may orchestrate SARS-CoV-2 infection and contribute to organ-specific tissue damage. In this respect, dissecting currently available knowledge of COVID-19 immunopathogenesis is crucially important, not only to improve our understanding of its pathophysiology but also to fuel the rationale of both novel and repurposed treatment modalities. Various immune-mediated pathways during SARS-CoV-2 infection are relevant in this context, which relate to innate immunity, adaptive immunity, and autoimmunity. Pathological findings in tissue specimens of patients with COVID-19 provide valuable information with regard to our understanding of pathophysiology as well as the development of evidence-based treatment regimens. This review provides an updated overview of the main pathological changes observed in COVID-19 within the most commonly affected organ systems, with special emphasis on immunopathology. Current management strategies for COVID-19 include supportive care and the use of repurposed or symptomatic drugs, such as dexamethasone, remdesivir, and anticoagulants. Ultimately, prevention is key to combat COVID-19, and this requires appropriate measures to attenuate its spread and, above all, the development and implementation of effective vaccines. © 2021 The Authors. The Journal of Pathology published by John Wiley & Sons, Ltd. on behalf of The Pathological Society of Great Britain and Ireland.
Keywords: acute respiratory distress syndrome (ARDS); angiotensin-converting enzyme 2 (ACE2); autoimmunity; coronavirus disease 2019 (COVID-19); diffuse alveolar damage (DAD); immunopathology; pathology; pathophysiology; severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2); treatment.
© 2021 The Authors. The Journal of Pathology published by John Wiley & Sons, Ltd. on behalf of The Pathological Society of Great Britain and Ireland.
Figures



References
-
- World Health Organization . Weekly epidemiological update – 27 January 2021. [Accessed 31 January 2021]. Available from: https://www.who.int/publications/m/item/weekly-epidemiological-update-27...
-
- Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID‐19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA 2020; 323 : 1239–1242. - PubMed
-
- Binkhorst M, Offringa A, Hoeven J. COVID‐19: comprehensive synopsis of suggested pathophysiological mechanisms and repurposed drugs. Preprints 2020; 10.20944/preprints202007.0108.v1. [Not peer reviewed]. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

