Anatomy and activity patterns in a multifunctional motor neuron and its surrounding circuits
- PMID: 33587033
- PMCID: PMC7954528
- DOI: 10.7554/eLife.61881
Anatomy and activity patterns in a multifunctional motor neuron and its surrounding circuits
Abstract
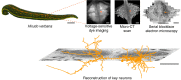
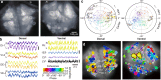
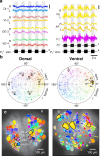
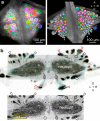
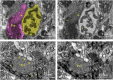
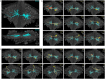
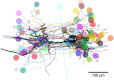
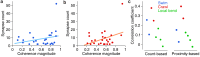
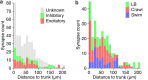
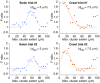
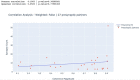
Dorsal Excitor motor neuron DE-3 in the medicinal leech plays three very different dynamical roles in three different behaviors. Without rewiring its anatomical connectivity, how can a motor neuron dynamically switch roles to play appropriate roles in various behaviors? We previously used voltage-sensitive dye imaging to record from DE-3 and most other neurons in the leech segmental ganglion during (fictive) swimming, crawling, and local-bend escape (Tomina and Wagenaar, 2017). Here, we repeated that experiment, then re-imaged the same ganglion using serial blockface electron microscopy and traced DE-3's processes. Further, we traced back the processes of DE-3's presynaptic partners to their respective somata. This allowed us to analyze the relationship between circuit anatomy and the activity patterns it sustains. We found that input synapses important for all the behaviors were widely distributed over DE-3's branches, yet that functional clusters were different during (fictive) swimming vs. crawling.
Keywords: Hirudo verbana; connectome; motor behaviors; neural circuit; neuroscience; serial blockface electron microscopy; voltage-sensitive dye imaging.
© 2021, Ashaber et al.
Conflict of interest statement
MA, YT, PK, EB, WK, ME, DW No competing interests declared
Figures









References
-
- Bates AS, Schlegel P, Roberts RJ, Drummond N, Tamimi IFM, Turnbull R, Zhao X, Marin EC, Popovici PD, Dhawan S, Jamasb A, Javier A, Li F, Rubin GM, Waddell S, Bock DD, Costa M, Jefferis G. Complete connectomic reconstruction of olfactory projection neurons in the fly brain. bioRxiv. 2020 doi: 10.1101/2020.01.19.911453. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

