Microbiome-based environmental monitoring of a dairy processing facility highlights the challenges associated with low microbial-load samples
- PMID: 33589631
- PMCID: PMC7884712
- DOI: 10.1038/s41538-021-00087-2
Microbiome-based environmental monitoring of a dairy processing facility highlights the challenges associated with low microbial-load samples
Abstract
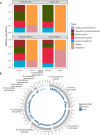
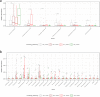
Efficient and accurate identification of microorganisms throughout the food chain can potentially allow the identification of sources of contamination and the timely implementation of control measures. High throughput DNA sequencing represents a potential means through which microbial monitoring can be enhanced. While Illumina sequencing platforms are most typically used, newer portable platforms, such as the Oxford Nanopore Technologies (ONT) MinION, offer the potential for rapid analysis of food chain microbiomes. Initial assessment of the ability of rapid MinION-based sequencing to identify microbes within a simple mock metagenomic mixture is performed. Subsequently, we compare the performance of both ONT and Illumina sequencing for environmental monitoring of an active food processing facility. Overall, ONT MinION sequencing provides accurate classification to species level, comparable to Illumina-derived outputs. However, while the MinION-based approach provides a means of easy library preparations and portability, the high concentrations of DNA needed is a limiting factor.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Gleeson D, O’Connell A, Jordan K. Review of potential sources and control of thermoduric bacteria in bulk-tank milk. Ir. J. Agric. Food Res. 2013;52:217–227.
-
- Fysun O, Kern H, Wilke B, Langowski H-C. Evaluation of factors influencing dairy biofilm formation in filling hoses of food-processing equipment. Food Bioprod. Process. 2019;113:39–48. doi: 10.1016/j.fbp.2018.10.009. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

