Identification of efficient prokaryotic cell-penetrating peptides with applications in bacterial biotechnology
- PMID: 33589718
- PMCID: PMC7884711
- DOI: 10.1038/s42003-021-01726-w
Identification of efficient prokaryotic cell-penetrating peptides with applications in bacterial biotechnology
Abstract
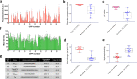
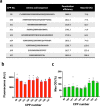
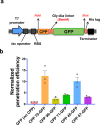
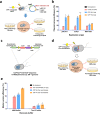
In bacterial biotechnology, instead of producing functional proteins from plasmids, it is often necessary to deliver functional proteins directly into live cells for genetic manipulation or physiological modification. We constructed a library of cell-penetrating peptides (CPPs) capable of delivering protein cargo into bacteria and developed an efficient delivery method for CPP-conjugated proteins. We screened the library for highly efficient CPPs with no significant cytotoxicity in Escherichia coli and developed a model for predicting the penetration efficiency of a query peptide, enabling the design of new and efficient CPPs. As a proof-of-concept, we used the CPPs for plasmid curing in E. coli and marker gene excision in Methylomonas sp. DH-1. In summary, we demonstrated the utility of CPPs in bacterial engineering. The use of CPPs would facilitate bacterial biotechnology such as genetic engineering, synthetic biology, metabolic engineering, and physiology studies.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

