Genome sequencing analysis identifies new loci associated with Lewy body dementia and provides insights into its genetic architecture
- PMID: 33589841
- PMCID: PMC7946812
- DOI: 10.1038/s41588-021-00785-3
Genome sequencing analysis identifies new loci associated with Lewy body dementia and provides insights into its genetic architecture
Abstract
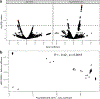
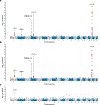
The genetic basis of Lewy body dementia (LBD) is not well understood. Here, we performed whole-genome sequencing in large cohorts of LBD cases and neurologically healthy controls to study the genetic architecture of this understudied form of dementia, and to generate a resource for the scientific community. Genome-wide association analysis identified five independent risk loci, whereas genome-wide gene-aggregation tests implicated mutations in the gene GBA. Genetic risk scores demonstrate that LBD shares risk profiles and pathways with Alzheimer's disease and Parkinson's disease, providing a deeper molecular understanding of the complex genetic architecture of this age-related neurodegenerative condition.
Figures



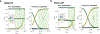



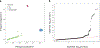




Comment in
-
Genetic insights into dementia disorders.Nat Rev Neurol. 2021 Apr;17(4):193. doi: 10.1038/s41582-021-00478-9. Nat Rev Neurol. 2021. PMID: 33649530 No abstract available.
References
-
- Meeus B, Theuns J & Van Broeckhoven C The genetics of dementia with Lewy bodies: what are we missing? Arch. Neurol. 69, 1113–1118 (2012). - PubMed
Methods-only References
-
- Emre M et al. Clinical diagnostic criteria for dementia associated with Parkinson’s disease. Mov. Disord 22, 1689–1707 (2007). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 AG013854/AG/NIA NIH HHS/United States
- K08 AG065463/AG/NIA NIH HHS/United States
- P30 AG066507/AG/NIA NIH HHS/United States
- G1100540/MRC_/Medical Research Council/United Kingdom
- U24 AG021886/AG/NIA NIH HHS/United States
- R01 AG017917/AG/NIA NIH HHS/United States
- ZIA AG000935/ImNIH/Intramural NIH HHS/United States
- G0900652/MRC_/Medical Research Council/United Kingdom
- G0502157/MRC_/Medical Research Council/United Kingdom
- P50 NS062684/NS/NINDS NIH HHS/United States
- U54 NS110435/NS/NINDS NIH HHS/United States
- U01 NS095736/NS/NINDS NIH HHS/United States
- R01 NS096740/NS/NINDS NIH HHS/United States
- U24 NS095871/NS/NINDS NIH HHS/United States
- P30 AG010161/AG/NIA NIH HHS/United States
- ZIA NS003034/ImNIH/Intramural NIH HHS/United States
- ZIA NS003033/ImNIH/Intramural NIH HHS/United States
- G0400074/MRC_/Medical Research Council/United Kingdom
- P50 NS072187/NS/NINDS NIH HHS/United States
- P30 AG072977/AG/NIA NIH HHS/United States
- U19 AG062418/AG/NIA NIH HHS/United States
- MR/L016397/1/MRC_/Medical Research Council/United Kingdom
- ZIA NS003154/ImNIH/Intramural NIH HHS/United States
- UL1 TR000040/TR/NCATS NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- MR/L023784/2/MRC_/Medical Research Council/United Kingdom
- UL1 TR001873/TR/NCATS NIH HHS/United States
- MR/N008324/1/MRC_/Medical Research Council/United Kingdom
- U01 NS100620/NS/NINDS NIH HHS/United States
- P50 NS071674/NS/NINDS NIH HHS/United States
- U54 NS100693/NS/NINDS NIH HHS/United States
- R01 NS115144/NS/NINDS NIH HHS/United States
- R01 AG015819/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

