Regulation of chromatin structure and function: insights into the histone chaperone FACT
- PMID: 33590780
- PMCID: PMC8018367
- DOI: 10.1080/15384101.2021.1881726
Regulation of chromatin structure and function: insights into the histone chaperone FACT
Abstract
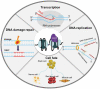
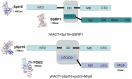
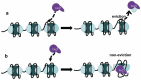
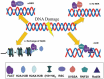
In eukaryotic cells, changes in chromatin accessibility are necessary for chromatin to maintain its highly dynamic nature at different times during the cell cycle. Histone chaperones interact with histones and regulate chromatin dynamics. Facilitates chromatin transcription (FACT) is an important histone chaperone that plays crucial roles during various cellular processes. Here, we analyze the structural characteristics of FACT, discuss how FACT regulates nucleosome/chromatin reorganization and summarize possible functions of FACT in transcription, replication, and DNA repair. The possible involvement of FACT in cell fate determination is also discussed.Abbreviations: FACT: facilitates chromatin transcription, Spt16: suppressor of Ty16, SSRP1: structure-specific recognition protein-1, NTD: N-terminal domain, DD: dimerization domain, MD: middle domain, CTD: C-terminus domain, IDD: internal intrinsically disordered domain, HMG: high mobility group, CID: C-terminal intrinsically disordered domain, Nhp6: non-histone chromosomal protein 6, RNAPII: RNA polymerase II, CK2: casein kinase 2, AID: acidic inner disorder, PIC: pre-initiation complex, IR: ionizing radiation, DDSB: DNA double-strand break, PARlation: poly ADP-ribosylation, BER: base-excision repair, UVSSA: UV-stimulated scaffold protein A, HR: homologous recombination, CAF-1: chromatin assembly factor 1, Asf1: anti-silencing factor 1, Rtt106: regulator of Ty1 transposition protein 106, H3K56ac: H3K56 acetylation, KD: knock down, SETD2: SET domain containing 2, H3K36me3: trimethylation of lysine36 in histone H3, H2Bub: H2B ubiquitination, iPSCs: induced pluripotent stem cells, ESC: embryonic stem cell, H3K4me3: trimethylation of lysine 4 on histone H3 protein subunit, CHD1: chromodomain protein.
Keywords: FACT; Histone chaperone; cell fate; chromatin dynamics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Luger K, Mader AW, Richmond RK, et al. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997 September18; 389(6648):251–260. - PubMed
-
- Talbert PB, Henikoff S.. Histone variants on the move: substrates for chromatin dynamics. Nat Rev Mol Cell Bio. 2017 Februaryl; 18(2):115–126. - PubMed
-
- Masliah-Planchon J, Bieche I, Guinebretiere JM, et al. SWI/SNF chromatin remodeling and human malignancies. Annu Rev Pathol. 2015;10(10):145–171. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
