long-read-tools.org: an interactive catalogue of analysis methods for long-read sequencing data
- PMID: 33590862
- PMCID: PMC7931822
- DOI: 10.1093/gigascience/giab003
long-read-tools.org: an interactive catalogue of analysis methods for long-read sequencing data
Abstract
Background: The data produced by long-read third-generation sequencers have unique characteristics compared to short-read sequencing data, often requiring tailored analysis tools for tasks ranging from quality control to downstream processing. The rapid growth in software that addresses these challenges for different genomics applications is difficult to keep track of, which makes it hard for users to choose the most appropriate tool for their analysis goal and for developers to identify areas of need and existing solutions to benchmark against.
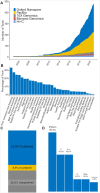
Findings: We describe the implementation of long-read-tools.org, an open-source database that organizes the rapidly expanding collection of long-read data analysis tools and allows its exploration through interactive browsing and filtering. The current database release contains 478 tools across 32 categories. Most tools are developed in Python, and the most frequent analysis tasks include base calling, de novo assembly, error correction, quality checking/filtering, and isoform detection, while long-read single-cell data analysis and transcriptomics are areas with the fewest tools available.
Conclusion: Continued growth in the application of long-read sequencing in genomics research positions the long-read-tools.org database as an essential resource that allows researchers to keep abreast of both established and emerging software to help guide the selection of the most relevant tool for their analysis needs.
Keywords: PacBio; data analysis; database; long-read sequencing; nanopore.
© The Author(s) 2021. Published by Oxford University Press GigaScience.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
-
- Mitsuhashi S, Matsumoto N. Long-read sequencing for rare human genetic diseases. J Hum Genet. 2020;65(1):11–9. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

