Genetic and epigenetic factors associated with increased severity of Covid-19
- PMID: 33590936
- PMCID: PMC8014716
- DOI: 10.1002/cbin.11572
Genetic and epigenetic factors associated with increased severity of Covid-19
Abstract
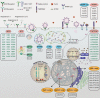
Since December 2019, a new form of severe acute respiratory syndrome (SARS) from a novel strain of coronavirus (SARS coronavirus 2 [SARS-CoV-2]) has been spreading worldwide. The disease caused by SARS-CoV-2 was named Covid-19 and declared as a pandemic by the World Health Organization in March 2020. Clinical symptoms of Covid-19 range from common cold to more severe disease defined as pneumonia, hypoxia, and severe respiratory distress. In the next stage, disease can become more critical with respiratory failure, sepsis, septic shock, and/or multiorgan failure. Outcomes of Covid-19 indicate large gaps between the male-female and the young-elder groups. Several theories have been proposed to explain variations, such as gender, age, comorbidity, and genetic factors. It is likely that mixture of genetic and nongenetic factors interplays between virus and host genetics and determines the severity of disease outcome. In this review, we aimed to summarize current literature in terms of potential host genetic and epigenetic factors that associated with increased severity of Covid-19. Several studies indicated that the genetic variants of the SARS-CoV-2 entry mechanism-related (angiotensin-converting enzymes, transmembrane serine protease-2, furin) and host innate immune response-related genes (interferons [IFNs], interleukins, toll-like receptors), and human leukocyte antigen, ABO, 3p21.31, and 9q34.2 loci are critical host determinants related to Covid-19 severity. Epigenetic mechanisms also affect Covid-19 outcomes by regulating IFN signaling, angiotensin-converting enzyme-2, and immunity-related genes that particularly escape from X chromosome inactivation. Enhanced understanding of host genetic and epigenetic factors and viral interactions of SARS-CoV-2 is critical for improved prognostic tools and innovative therapeutics.
Keywords: Covid-19; SARS-CoV-2; disease severity; epigenetic; genetic predisposition; host genetic factors.
© 2021 International Federation for Cell Biology.
Figures
References
-
- Agostini, S. , Mancuso, R. , Guerini, F. R. , D'Alfonso, S. , Agliardi, C. , Hernis, A. , Zanzottera, M. , Barizzone, N. , Leone, M. A. , Caputo, D. , Rovaris, M. , & Clerici, M. (2018). HLA alleles modulate EBV viral load in multiple sclerosis. Journal of Translational Medicine, 16(1), 80. 10.1186/s12967-018-1450-6 - DOI - PMC - PubMed
-
- Arts, R. J. W. , Moorlag, S. J. C. F. M. , Novakovic, B. , Li, Y. , Wang, S. Y. , Oosting, M. , Kumar, V. , Xavier, R. J. , Wijmenga, C. , Joosten, L. A. B. , Reusken, C. B. E. M. , Benn, C. S. , Aaby, P. , Koopmans, M. P. , Stunnenberg, H. G. , van Crevel, R. , & Netea, M. G. (2018). BCG vaccination protects against experimental viral infection in humans through the induction of cytokines associated with trained immunity. Cell Host & Microbe, 23(1), 89–100. 10.1016/j.chom.2017.12.010 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

