Junction plakoglobin regulates and destabilizes HIF2α to inhibit tumorigenesis of renal cell carcinoma
- PMID: 33591636
- PMCID: PMC8045910
- DOI: 10.1002/cac2.12142
Junction plakoglobin regulates and destabilizes HIF2α to inhibit tumorigenesis of renal cell carcinoma
Abstract
Background: Increased hypoxia-inducible factor 2α (HIF2α) activation is a common event in clear cell renal cell carcinoma (ccRCC) progression. However, the function and underlying mechanism of HIF2α in ccRCC remains uninvestigated. We conducted this study to access the potential link between junction plakoglobin (JUP) and HIF2α in ccRCC.
Methods: Affinity purification and mass spectrometry (AP-MS) screening, glutathione-s-transferase (GST) pull-down and co-immunoprecipitation (Co-IP) assays were performed to detect the interacting proteins of HIF2α. Quantitative PCR (qPCR) and Western blotting were used to detect the expression of JUP in human ccRCC samples. Luciferase reporter assays, chromatin immunoprecipitation (ChIP), cycloheximide chase assays, and ubiquitination assays were conducted to explore the regulation of JUP on the activity of HIF2α. Cell Counting Kit-8 (CCK-8) assays, colony formation assays, transwell assays, and xenograft tumor assays were performed to investigate the effect of JUP knockdown or overexpression on the tumorigenicity of renal cancer cells.
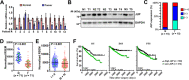
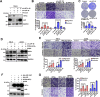
Results: We identified JUP as a novel HIF2α-binding partner and revealed an important role of JUP in recruiting von Hippel-Lindau (VHL) and histone deacetylases 1/2 (HDAC1/2) to HIF2α to regulate its stability and transactivation. JUP knockdown promoted and overexpression suppressed the tumorigenicity of renal cell carcinoma in vitro and in vivo. Importantly, the low expression of JUP was found in clinical ccRCC samples and correlated with enhanced hypoxia scores and poor treatment outcomes.
Conclusion: Taken together, these data support a role of JUP in modulating HIF2α signaling during ccRCC progression and identify JUP as a potential therapeutic target.
Keywords: hypoxia-inducible factor 2α; junction plakoglobin; renal cell carcinoma; transcriptional activity; ubiquitination.
© 2021 The Authors. Cancer Communications published by John Wiley & Sons Australia, Ltd. on behalf of Sun Yat-sen University Cancer Center.
Conflict of interest statement
No potential conflicts of interest were disclosed.
Figures





References
-
- Capitanio U, Montorsi F. Renal cancer. Lancet. 2016;387(10021):894–906. - PubMed
-
- Maxwell PH, Wiesener MS, Chang G‐W, Clifford SC, Vaux EC, Cockman ME, et al. The tumour suppressor protein VHL targets hypoxia‐inducible factors for oxygen‐dependent proteolysis. Nature. 1999;399(6733):271–5. - PubMed
-
- Zou JX, Chen K. Roles and molecular mechanisms of hypoxia‐inducible factors in renal cell carcinoma. Yi Chuan. 2018;40(5):341–56. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

