Imd pathway-specific immune assays reveal NF-κB stimulation by viral RNA PAMPs in Aedes aegypti Aag2 cells
- PMID: 33591970
- PMCID: PMC7909628
- DOI: 10.1371/journal.pntd.0008524
Imd pathway-specific immune assays reveal NF-κB stimulation by viral RNA PAMPs in Aedes aegypti Aag2 cells
Abstract
Background: The mosquito Aedes aegypti is a major vector for the arthropod-borne viruses (arboviruses) chikungunya, dengue, yellow fever and Zika viruses. Vector immune responses pose a major barrier to arboviral transmission, and transgenic insects with altered immunity have been proposed as tools for reducing the global public health impact of arboviral diseases. However, a better understanding of virus-immune interactions is needed to progress the development of such transgenic insects. Although the NF-κB-regulated Toll and 'immunodeficiency' (Imd) pathways are increasingly thought to be antiviral, relevant pattern recognition receptors (PRRs) and pathogen-associated molecular patterns (PAMPs) remain poorly characterised in A. aegypti.
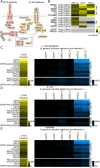
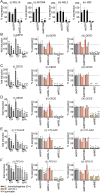
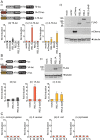
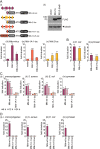
Methodology/principle findings: We developed novel RT-qPCR and luciferase reporter assays to measure induction of the Toll and Imd pathways in the commonly used A. aegypti-derived Aag2 cell line. We thus determined that the Toll pathway is not inducible by exogenous stimulation with bacterial, viral or fungal stimuli in Aag2 cells under our experimental conditions. We used our Imd pathway-specific assays to demonstrate that the viral dsRNA mimic poly(I:C) is sensed by the Imd pathway, likely through intracellular and extracellular PRRs. The Imd pathway was also induced during infection with the model insect-specific virus cricket paralysis virus (CrPV).
Conclusions/significance: Our demonstration that a general PAMP shared by many arboviruses is sensed by the Imd pathway paves the way for future studies to determine how viral RNA is sensed by mosquito PRRs at a molecular level. Our data also suggest that studies measuring inducible immune pathway activation through antimicrobial peptide (AMP) expression in Aag2 cells should be interpreted cautiously given that the Toll pathway is not responsive under all experimental conditions. With no antiviral therapies and few effective vaccines available to treat arboviral diseases, our findings provide new insights relevant to the development of transgenic mosquitoes as a means of reducing arbovirus transmission.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Aedes aegypti (Aag2)-derived clonal mosquito cell lines reveal the effects of pre-existing persistent infection with the insect-specific bunyavirus Phasi Charoen-like virus on arbovirus replication.PLoS Negl Trop Dis. 2019 Nov 6;13(11):e0007346. doi: 10.1371/journal.pntd.0007346. eCollection 2019 Nov. PLoS Negl Trop Dis. 2019. PMID: 31693659 Free PMC article.
-
Cell-Fusing Agent Virus Reduces Arbovirus Dissemination in Aedes aegypti Mosquitoes In Vivo.J Virol. 2019 Aug 28;93(18):e00705-19. doi: 10.1128/JVI.00705-19. Print 2019 Sep 15. J Virol. 2019. PMID: 31243123 Free PMC article.
-
The Aedes aegypti Domino Ortholog p400 Regulates Antiviral Exogenous Small Interfering RNA Pathway Activity and ago-2 Expression.mSphere. 2020 Apr 8;5(2):e00081-20. doi: 10.1128/mSphere.00081-20. mSphere. 2020. PMID: 32269152 Free PMC article.
-
Antiviral Compounds for Blocking Arboviral Transmission in Mosquitoes.Viruses. 2021 Jan 14;13(1):108. doi: 10.3390/v13010108. Viruses. 2021. PMID: 33466915 Free PMC article. Review.
-
The immune strategies of mosquito Aedes aegypti against microbial infection.Dev Comp Immunol. 2018 Jun;83:12-21. doi: 10.1016/j.dci.2017.12.001. Epub 2017 Dec 5. Dev Comp Immunol. 2018. PMID: 29217264 Review.
Cited by
-
Transcriptional response of Wolbachia-transinfected Aedes aegypti mosquito cells to dengue virus at early stages of infection.J Gen Virol. 2022 Jan;103(1):001694. doi: 10.1099/jgv.0.001694. J Gen Virol. 2022. PMID: 35006065 Free PMC article.
-
Cell Line Platforms Support Research into Arthropod Immunity.Insects. 2021 Aug 17;12(8):738. doi: 10.3390/insects12080738. Insects. 2021. PMID: 34442304 Free PMC article. Review.
-
Rift Valley Fever Virus Primes Immune Responses in Aedes aegypti Cells.Pathogens. 2023 Apr 6;12(4):563. doi: 10.3390/pathogens12040563. Pathogens. 2023. PMID: 37111448 Free PMC article.
-
Dengue Virus-2 Infection Affects Fecundity and Elicits Specific Transcriptional Changes in the Ovaries of Aedes aegypti Mosquitoes.Front Microbiol. 2022 Jun 23;13:886787. doi: 10.3389/fmicb.2022.886787. eCollection 2022. Front Microbiol. 2022. PMID: 35814655 Free PMC article.
-
Current Status of Immune Deficiency Pathway in Tenebrio molitor Innate Immunity.Front Immunol. 2022 Jul 4;13:906192. doi: 10.3389/fimmu.2022.906192. eCollection 2022. Front Immunol. 2022. PMID: 35860244 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

