Evaluation of two automated low-cost RNA extraction protocols for SARS-CoV-2 detection
- PMID: 33591986
- PMCID: PMC7886139
- DOI: 10.1371/journal.pone.0246302
Evaluation of two automated low-cost RNA extraction protocols for SARS-CoV-2 detection
Abstract
Background: Two automatable in-house protocols for high-troughput RNA extraction from nasopharyngeal swabs for SARS-CoV-2 detection have been evaluated.
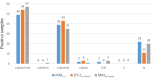
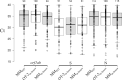
Methods: One hundred forty one SARS-CoV-2 positive samples were collected during a period of 10-days. In-house protocols were based on extraction with magnetic beads and designed to be used with either the Opentrons OT-2 (OT-2in-house) liquid handling robot or the MagMAXTM Express-96 system (MMin-house). Both protocols were tested in parallel with a commercial kit that uses the MagMAXTM system (MMkit). Nucleic acid extraction efficiencies were calculated from a SARS-CoV-2 DNA positive control.
Results: No significant differences were found between both in-house protocols and the commercial kit in their performance to detect positive samples. The MMkit was the most efficient although the MMin-house presented, in average, lower Cts than the other two. In-house protocols allowed to save between 350€ and 400€ for every 96 extracted samples compared to the commercial kit.
Conclusion: The protocols described harness the use of easily available reagents and an open-source liquid handling system and are suitable for SARS-CoV-2 detection in high throughput facilities.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

